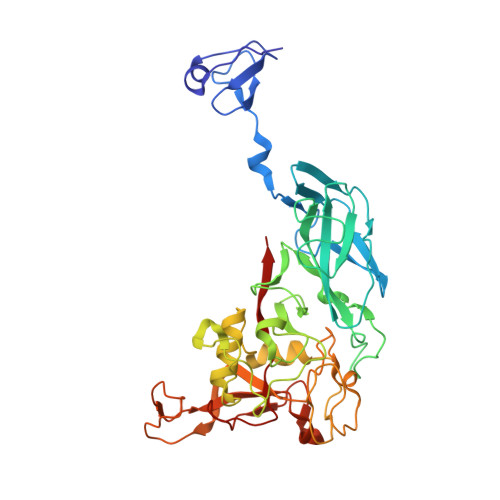
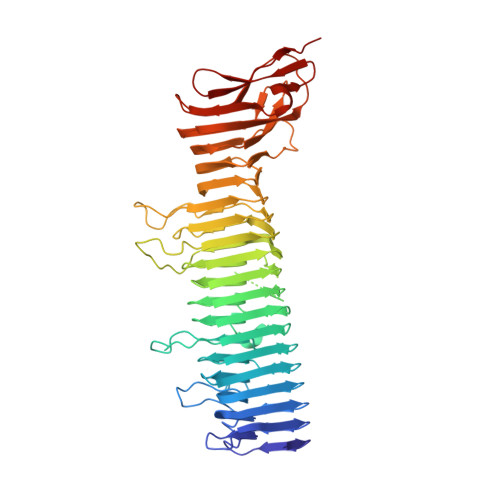
Selective Ligand Recognition by a Diversity-Generating Retroelement Variable Protein
Miller, J.L., Le Coq, J., Hodes, A., Barbalat, R., Miller, J.F., Ghosh, P.(2008) PLoS Biol 6: e131-e131
- PubMed: 18532877
- DOI: https://doi.org/10.1371/journal.pbio.0060131
- Primary Citation of Related Structures:
2IOU - PubMed Abstract:
Diversity-generating retroelements (DGRs) recognize novel ligands through massive protein sequence variation, a property shared uniquely with the adaptive immune response. Little is known about how recognition is achieved by DGR variable proteins. Here, we present the structure of the Bordetella bacteriophage DGR variable protein major tropism determinant (Mtd) bound to the receptor pertactin, revealing remarkable adaptability in the static binding sites of Mtd. Despite large dissimilarities in ligand binding mode, principles underlying selective recognition were strikingly conserved between Mtd and immunoreceptors. Central to this was the differential amplification of binding strengths by avidity (i.e., multivalency), which not only relaxed the demand for optimal complementarity between Mtd and pertactin but also enhanced distinctions among binding events to provide selectivity. A quantitatively similar balance between complementarity and avidity was observed for Bordetella bacteriophage DGR as occurs in the immune system, suggesting that variable repertoires operate under a narrow set of conditions to recognize novel ligands.
- Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, California, United States of America.
Organizational Affiliation: