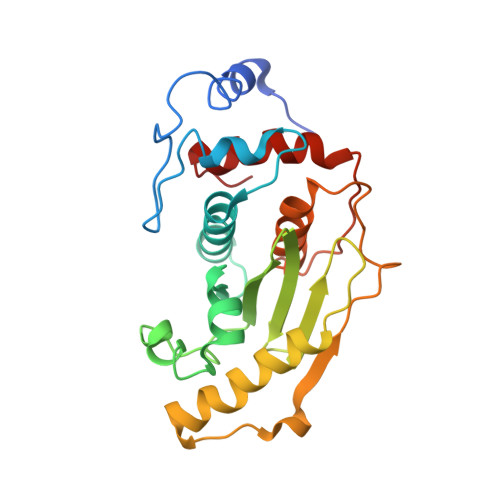
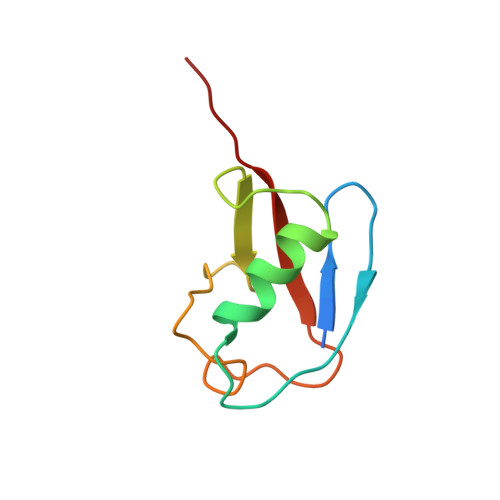
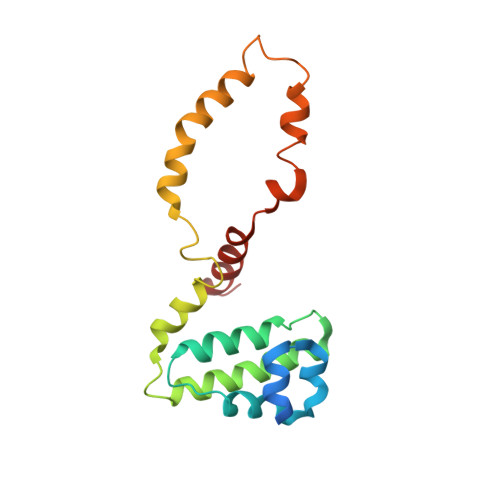
Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates.
Reverter, D., Lima, C.D.(2006) Nat Struct Mol Biol 13: 1060-1068
- PubMed: 17099700
- DOI: https://doi.org/10.1038/nsmb1168
- Primary Citation of Related Structures:
2IO0, 2IO1, 2IO2, 2IO3 - PubMed Abstract:
SUMO processing and deconjugation are essential proteolytic activities for nuclear metabolism and cell-cycle progression in yeast and higher eukaryotes. To elucidate the mechanisms used during substrate lysine deconjugation, SUMO isoform processing and SUMO isoform interactions, X-ray structures were determined for a catalytically inert SENP2 protease domain in complex with conjugated RanGAP1-SUMO-1 or RanGAP1-SUMO-2, or in complex with SUMO-2 or SUMO-3 precursors. Common features within the active site include a 90 degrees kink proximal to the scissile bond that forces C-terminal amino acid residues or the lysine side chain toward a protease surface that appears optimized for lysine deconjugation. Analysis of this surface reveals SENP2 residues, particularly Met497, that mediate, and in some instances reverse, in vitro substrate specificity. Mutational analysis and biochemistry provide a mechanism for SENP2 substrate preferences that explains why SENP2 catalyzes SUMO deconjugation more efficiently than processing.
- Structural Biology Program, Sloan-Kettering Institute, New York, New York 10021, USA.
Organizational Affiliation: