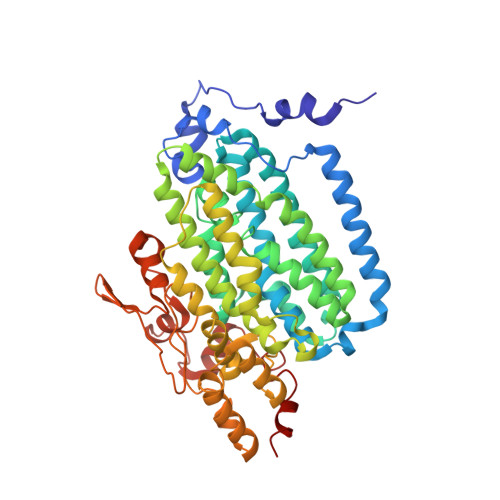
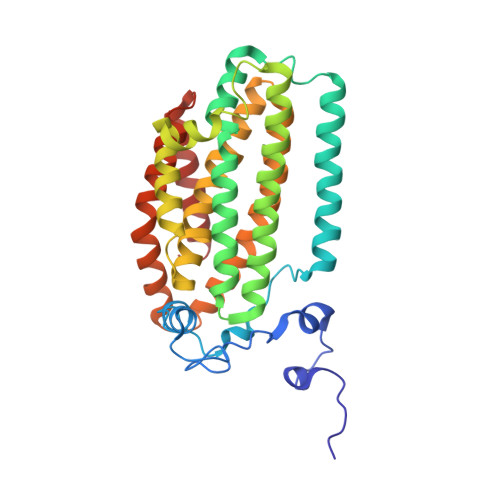
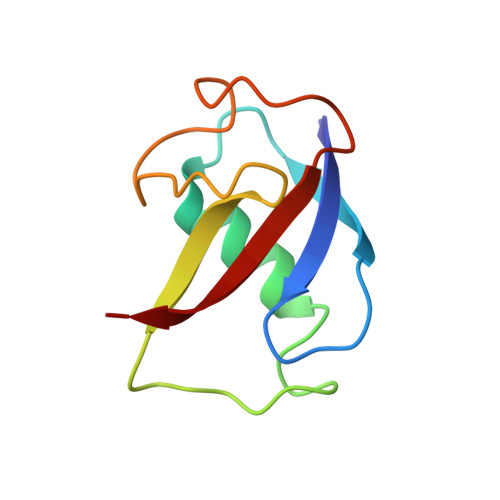
X-ray crystal structures of manganese(II)-reconstituted and native toluene/o-xylene monooxygenase hydroxylase reveal rotamer shifts in conserved residues and an enhanced view of the protein interior.
McCormick, M.S., Sazinsky, M.H., Condon, K.L., Lippard, S.J.(2006) J Am Chem Soc 128: 15108-15110
- PubMed: 17117860
- DOI: https://doi.org/10.1021/ja064837r
- Primary Citation of Related Structures:
2INC, 2IND - PubMed Abstract:
We report the X-ray crystal structures of native and manganese(II)-reconstituted toluene/o-xylene monooxygenase hydroxylase (ToMOH) from Pseudomonas stutzeri OX1 to 1.85 and 2.20 A resolution, respectively. The structures reveal that reduction of the dimetallic active site is accompanied by a carboxylate shift and alteration of the coordination environment for dioxygen binding and activation. A rotamer shift in a strategically placed asparagine 202 accompanies dimetallic center reduction and is proposed to influence protein component interactions. This rotamer shift is conserved between ToMOH and the corresponding residue in methane monooxygenase hydroxylase (MMOH). Previously unidentified hydrophobic pockets similar to those present in MMOH are assigned.
- Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA.
Organizational Affiliation: