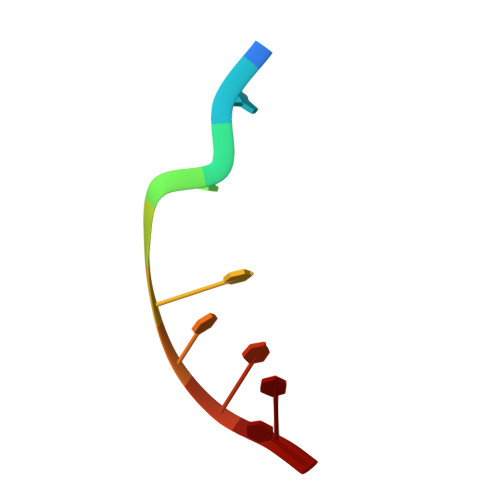
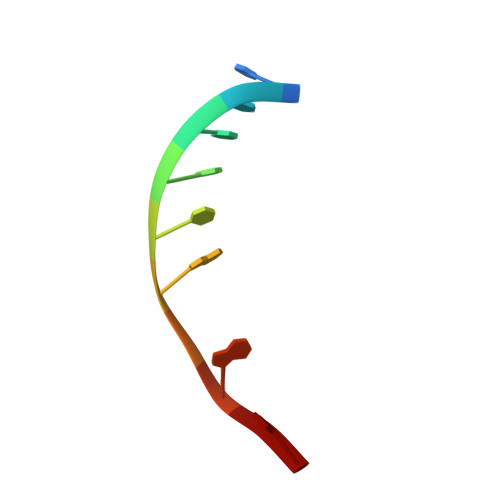
Toward a Designed, Functioning Genetic System with Expanded-Size Base Pairs: Solution Structure of the Eight-Base xDNA Double Helix.
Lynch, S.R., Liu, H., Gao, J., Kool, E.T.(2006) J Am Chem Soc 128: 14704-14711
- PubMed: 17090058
- DOI: https://doi.org/10.1021/ja065606n
- Primary Citation of Related Structures:
2ICZ - PubMed Abstract:
We describe the NMR-derived solution structure of the double-helical form of a designed eight-base genetic pairing system, termed xDNA. The benzo-homologous xDNA design contains base pairs that are wider than natural DNA pairs by ca. 2.4 A (the width of a benzene ring). The eight component bases of this xDNA helix are A, C, G, T, xA, xT, xC, and xG. The structure was solved in aqueous buffer using 1D and 2D NMR methods combined with restrained molecular dynamics. The data show that the decamer duplex is right-handed and antiparallel, and hydrogen-bonded in a way analogous to that of Watson-Crick DNA. The sugar-phosphate backbone adopts a regular conformation similar to that of B-form DNA, with small dihedral adjustments due to the larger circumference of the helix. The grooves are much wider and more shallow than those of B-form DNA, and the helix turn is slower, with ca. 12 base pairs per 360 degrees turn. There is an extensive intra- and interstrand base stacking surface area, providing an explanation for the greater stability of xDNA relative to natural DNA. There is also evidence for greater motion in this structure compared to a previous two-base-expanded helix; possible chemical and structural reasons for this are discussed. The results confirm paired self-assembly of the designed xDNA system. This suggests the possibility that other genetic system structures besides the natural one might be functional in encoding information and transferring it to new complementary strands.
- Department of Chemistry, Stanford University, Stanford, California 94305-5080, USA.
Organizational Affiliation: