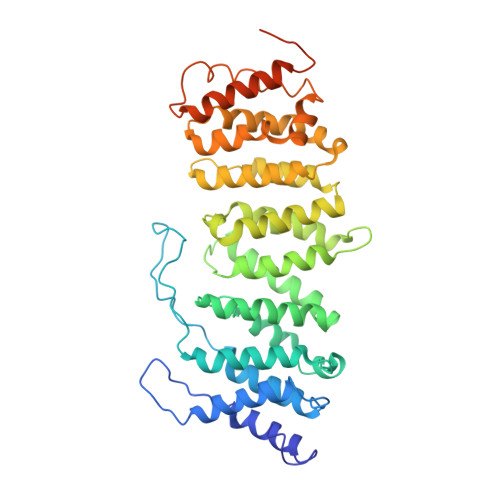
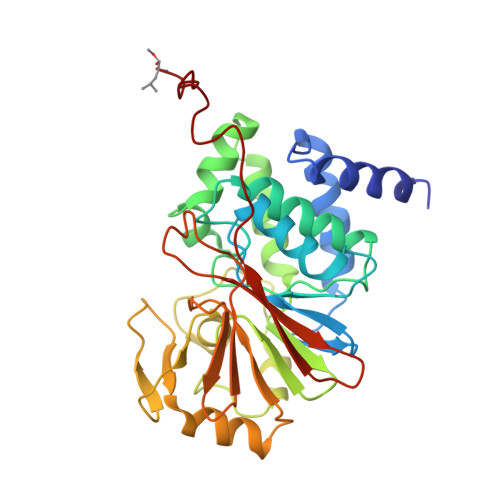
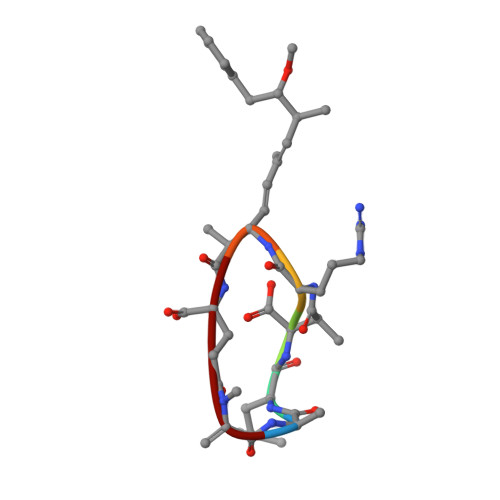
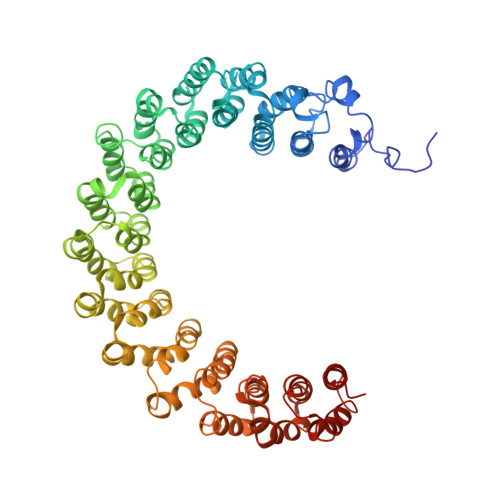Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme.
Cho, U.S., Xu, W.(2007) Nature 445: 53-57
- PubMed: 17086192
- DOI: https://doi.org/10.1038/nature05351
- Primary Citation of Related Structures:
2IAE - PubMed Abstract:
Protein phosphatase 2A (PP2A) is a principal Ser/Thr phosphatase, the deregulation of which is associated with multiple human cancers, Alzheimer's disease and increased susceptibility to pathogen infections. How PP2A is structurally organized and functionally regulated remains unclear. Here we report the crystal structure of an AB'C heterotrimeric PP2A holoenzyme. The structure reveals that the HEAT repeats of the scaffold A subunit form a horseshoe-shaped fold, holding the catalytic C and regulatory B' subunits together on the same side. The regulatory B' subunit forms pseudo-HEAT repeats and interacts with the C subunit near the active site, thereby defining substrate specificity. The methylated carboxy-terminal tail of the C subunit interacts with a highly negatively charged region at the interface between A and B' subunits, suggesting that the C-terminal carboxyl methylation of the C subunit promotes B' subunit recruitment by neutralizing charge repulsion. Together, our structural results establish a crucial foundation for understanding PP2A assembly, substrate recruitment and regulation.
- Department of Biological Structure, University of Washington, Seattle, Washington 98195, USA.
Organizational Affiliation: