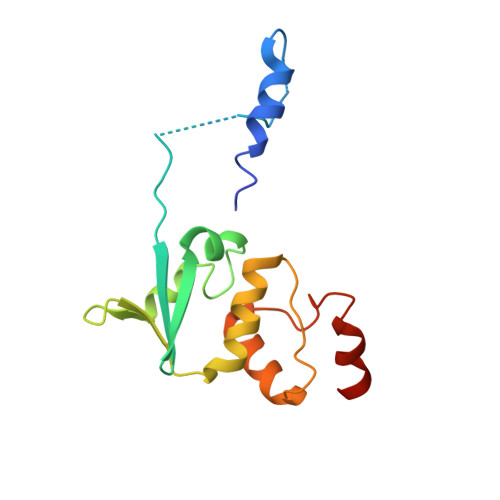
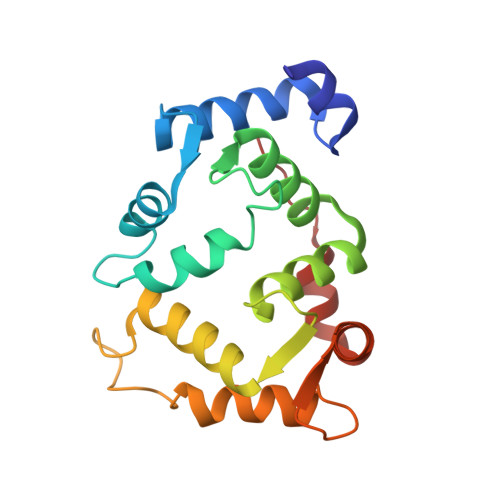
Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer
Pioletti, M., Findeisen, F., Hura, G.L., Minor Jr., D.L.(2006) Nat Struct Mol Biol 13: 987-995
- PubMed: 17057713
- DOI: https://doi.org/10.1038/nsmb1164
- Primary Citation of Related Structures:
2I2R - PubMed Abstract:
Brain I(A) and cardiac I(to) currents arise from complexes containing Kv4 voltage-gated potassium channels and cytoplasmic calcium-sensor proteins (KChIPs). Here, we present X-ray crystallographic and small-angle X-ray scattering data that show that the KChIP1-Kv4.3 N-terminal cytoplasmic domain complex is a cross-shaped octamer bearing two principal interaction sites. Site 1 comprises interactions between a unique Kv4 channel N-terminal hydrophobic segment and a hydrophobic pocket formed by displacement of the KChIP H10 helix. Site 2 comprises interactions between a T1 assembly domain loop and the KChIP H2 helix. Functional and biochemical studies indicate that site 1 influences channel trafficking, whereas site 2 affects channel gating, and that calcium binding is intimately linked to KChIP folding and complex formation. Together, the data resolve how Kv4 channels and KChIPs interact and provide a framework for understanding how KChIPs modulate Kv4 function.
- Cardiovascular Research Institute, Department of Biochemistry, California Institute for Quantitative Biomedical Research, University of California, San Francisco, California 94143-2532, USA.
Organizational Affiliation: