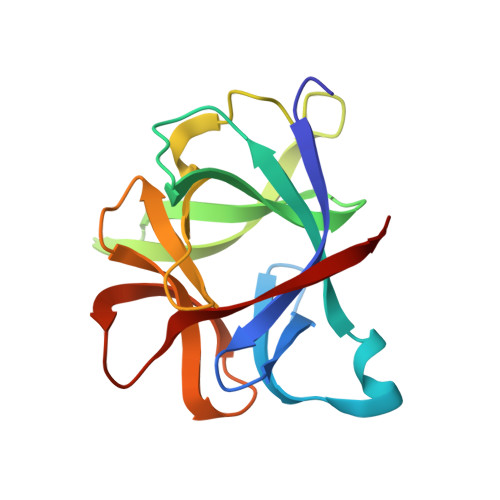
Crystallographic refinement of interleukin 1 beta at 2.0 A resolution.
Priestle, J.P., Schar, H.P., Grutter, M.G.(1989) Proc Natl Acad Sci U S A 86: 9667-9671
- PubMed: 2602367
- DOI: https://doi.org/10.1073/pnas.86.24.9667
- Primary Citation of Related Structures:
2I1B - PubMed Abstract:
The structure of human recombinant interleukin 1 beta (IL-1 beta) has been refined by a restrained least-squares method to a crystallographic R factor of 17.2% to 2.0 A resolution. One-hundred sixty-eight solvent molecules have been located, and isotropic temperature factors for each atom have been refined. The overall structure is composed of 12 beta-strands that can best be described as forming the four triangular faces of a tetrahedron with hydrogen bonding resembling normal antiparallel beta-sheets only at the vertices. The interior of this tetrahedron is filled by hydrophobic side chains. Analysis of sequence alignments with IL-1 beta from other mammalian species shows the interior to be very well conserved with the exterior residues markedly less so. There does not appear to be a clustering of invariant amino acid side chains on the surface of the molecule, suggesting an area of interaction with the IL-1 receptor. Comparison of the IL-1 beta structure with IL-1 alpha sequences indicates that IL-1 alpha probably has a similar overall folding as IL-1 beta but binds to the receptor in a different fashion. The three-dimensional structure of the IL-1 beta is analyzed in light of what has been suggested by previously published work on mutants and fragments of the molecule.
- Pharmaceuticals Research Division, CIBA-Geigy Ltd., Basel, Switzerland.
Organizational Affiliation: