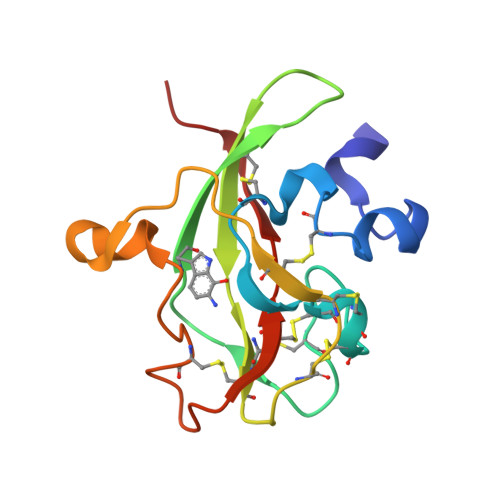
New insights into the reductive half-reaction mechanism of aromatic amine dehydrogenase revealed by reaction with carbinolamine substrates.
Roujeinikova, A., Hothi, P., Masgrau, L., Sutcliffe, M.J., Scrutton, N.S., Leys, D.(2007) J Biological Chem 282: 23766-23777
- PubMed: 17475620
- DOI: https://doi.org/10.1074/jbc.M700677200
- Primary Citation of Related Structures:
2I0R, 2I0S, 2I0T, 2OIZ, 2OJY, 2OK4, 2OK6 - PubMed Abstract:
Aromatic amine dehydrogenase uses a tryptophan tryptophylquinone (TTQ) cofactor to oxidatively deaminate primary aromatic amines. In the reductive half-reaction, a proton is transferred from the substrate C1 to betaAsp-128 O-2, in a reaction that proceeds by H-tunneling. Using solution studies, kinetic crystallography, and computational simulation we show that the mechanism of oxidation of aromatic carbinolamines is similar to amine oxidation, but that carbinolamine oxidation occurs at a substantially reduced rate. This has enabled us to determine for the first time the structure of the intermediate prior to the H-transfer/reduction step. The proton-betaAsp-128 O-2 distance is approximately 3.7A, in contrast to the distance of approximately 2.7A predicted for the intermediate formed with the corresponding primary amine substrate. This difference of approximately 1.0 A is due to an unexpected conformation of the substrate moiety, which is supported by molecular dynamic simulations and reflected in the approximately 10(7)-fold slower TTQ reduction rate with phenylaminoethanol compared with that with primary amines. A water molecule is observed near TTQ C-6 and is likely derived from the collapse of the preceding carbinolamine TTQ-adduct. We suggest this water molecule is involved in consecutive proton transfers following TTQ reduction, and is ultimately repositioned near the TTQ O-7 concomitant with protein rearrangement. For all carbinolamines tested, highly stable amide-TTQ adducts are formed following proton abstraction and TTQ reduction. Slow hydrolysis of the amide occurs after, rather than prior to, TTQ oxidation and leads ultimately to a carboxylic acid product.
- Manchester Interdisciplinary Biocenter, University of Manchester, Manchester M17DN, United Kingdom.
Organizational Affiliation: