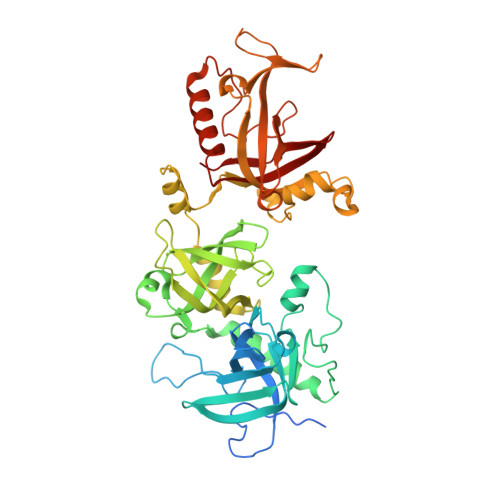
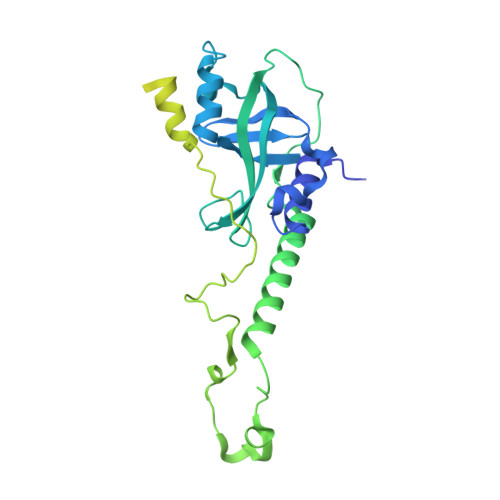
Structural reorganization and the cooperative binding of single-stranded telomere DNA in Sterkiella nova.
Buczek, P., Horvath, M.P.(2006) J Biological Chem 281: 40124-40134
- PubMed: 17082188
- DOI: https://doi.org/10.1074/jbc.M607749200
- Primary Citation of Related Structures:
2I0Q - PubMed Abstract:
In Sterkiella nova, alpha and beta telomere proteins bind cooperatively with single-stranded DNA to form a ternary alpha.beta.DNA complex. Association of telomere protein subunits is DNA-dependent, and alpha-beta association enhances DNA affinity. To further understand the molecular basis for binding cooperativity, we characterized several possible stepwise assembly pathways using isothermal titration calorimetry. In one path, alpha and DNA first form a stable alpha.DNA complex followed by the addition of beta in a second step. Binding energy accumulates with nearly equal free energy of association for each of these steps. Heat capacity is nonetheless dramatically different, with DeltaCp = -305 +/- 3 cal mol(-1) K(-1) for alpha binding with DNA and DeltaCp = -2010 +/- 20 cal mol(-1) K(-1) for the addition of beta to complete the alpha.beta.DNA complex. By examining alternate routes including titration of single-stranded DNA with a preformed alpha.beta complex, a significant portion of binding energy and heat capacity could be assigned to structural reorganization involving protein-protein interactions and repositioning of the DNA. Structural reorganization probably affords a mechanism to regulate high affinity binding of telomere single-stranded DNA with important implications for telomere biology. Regulation of telomere complex dissociation is thought to involve post-translational modifications in the lysine-rich C-terminal portion of beta. We observed no difference in binding energetics or crystal structure when comparing complexes prepared with full-length beta or a C-terminally truncated form, supporting interesting parallels between the intrinsically disordered regions of histones and this portion of beta.
- Department of Biology, University of Utah, Salt Lake City, Utah 84112, USA.
Organizational Affiliation: