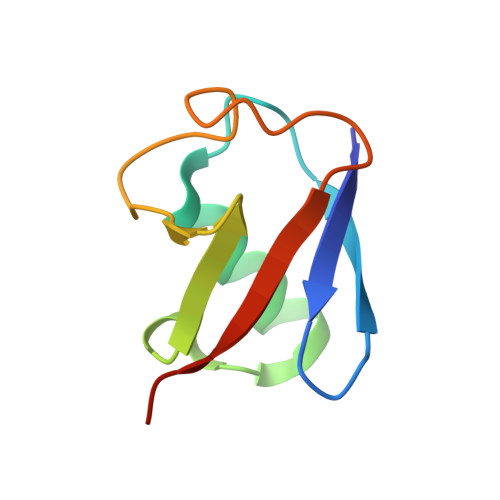
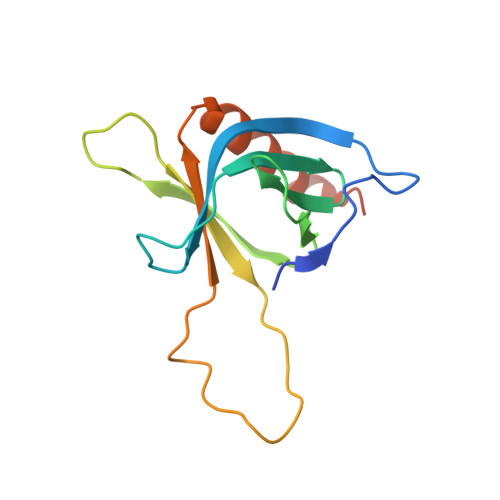
Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain.
Alam, S.L., Langelier, C., Whitby, F.G., Koirala, S., Robinson, H., Hill, C.P., Sundquist, W.I.(2006) Nat Struct Mol Biol 13: 1029-1030
- PubMed: 17057716
- DOI: https://doi.org/10.1038/nsmb1160
- Primary Citation of Related Structures:
2HTH - PubMed Abstract:
The ESCRT-I and ESCRT-II complexes help sort ubiquitinated proteins into vesicles that accumulate within multivesicular bodies (MVBs). Crystallographic and biochemical analyses reveal that the GLUE domain of the human ESCRT-II EAP45 (also called VPS36) subunit is a split pleckstrin-homology domain that binds ubiquitin along one edge of the beta-sandwich. The structure suggests how human ESCRT-II can couple recognition of ubiquitinated cargoes and endosomal phospholipids during MVB protein sorting.
- Department of Biochemistry, University of Utah, Salt Lake City, Utah 84112-5650, USA.
Organizational Affiliation: