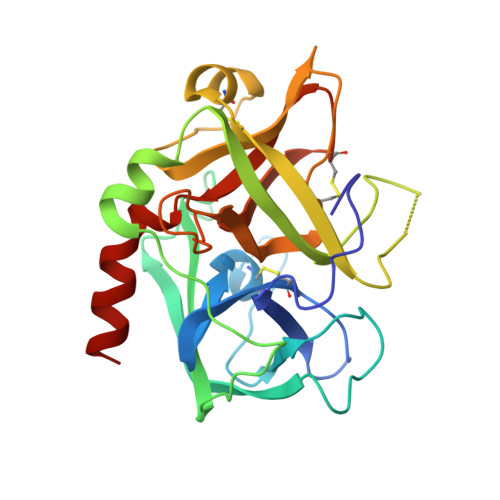
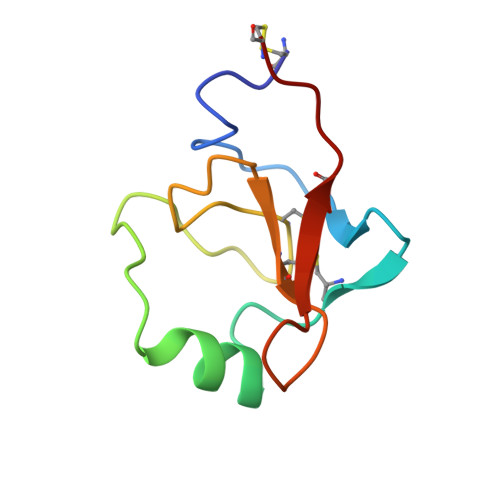
Structures of the noncovalent complexes of human and bovine prothrombin fragment 2 with human PPACK-thrombin.
Arni, R.K., Padmanabhan, K., Padmanabhan, K.P., Wu, T.P., Tulinsky, A.(1993) Biochemistry 32: 4727-4737
- PubMed: 8387813
- DOI: https://doi.org/10.1021/bi00069a006
- Primary Citation of Related Structures:
2HPP, 2HPQ - PubMed Abstract:
Both human and bovine prothrombin fragment 2 (the second kringle) have been cocrystallized separately with human PPACK (D-Phe-Pro-Arg)-thrombin, and the structures of these noncovalent complexes have been determined and refined (R = 0.155 and 0.157, respectively) at 3.3-A resolution using X-ray crystallographic methods. The kringles interact with thrombin at a site that has previously been proposed to be the heparin binding region. The latter is a highly electropositive surface near the C-terminal helix of thrombin abundant in arginine and lysine residues. These form salt bridges with acidic side chains of kringle 2. Somewhat unexpectedly, the negative groups of the kringle correspond to an enlarged anionic center of the lysine binding site of lysine binding kringles such as plasminogens K1 and K4 and TPA K2. The anionic motif is DGDEE in prothrombin kringle 2. The corresponding cationic center of the lysine binding site region has an unfavorable Arg70Asp substitution, but Lys35 is conserved. However, the folding of fragment 2 is different from that of prothrombin kringle 1 and other kringles: the second outer loop possesses a distorted two-turn helix, and the hairpin beta-turn of the second inner loop pivots at Val64 and Asp70 by 60 degrees. Lys35 is located on a turn of the helix, which causes it to project into solvent space in the fragment 2-thrombin complex, thereby devastating any vestige of the cationic center of the lysine binding site. Since fragment 2 has not been reported to bind lysine, it most likely has a different inherent folding conformation for the second outer loop, as has also been observed to be the case with TPA K2 and the urokinase kringle. The movement of the Val64-Asp70 beta-turn is most likely a conformational change accompanying complexation, which reveals a new heretofore unsuspected flexibility in kringles. The fragment 2-thrombin complex is only the second cassette module-catalytic domain structure to be determined for a multidomain blood protein and only the third domain-domain interaction to be described among such proteins, the others being factor Xa without a Gla domain and Ca2+ prothrombin fragment 1 with a Gla domain and a kringle.
- Department of Chemistry, Michigan State University, East Lansing 48824-1322.
Organizational Affiliation: