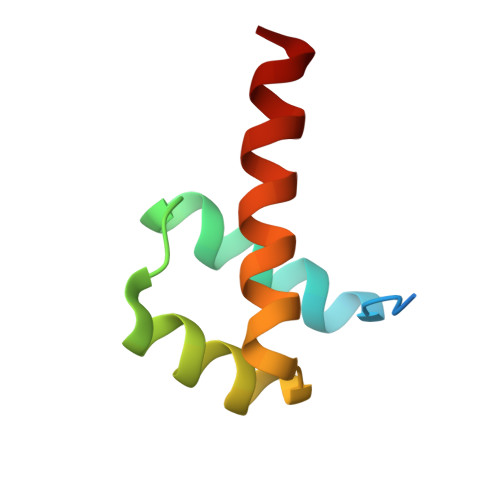
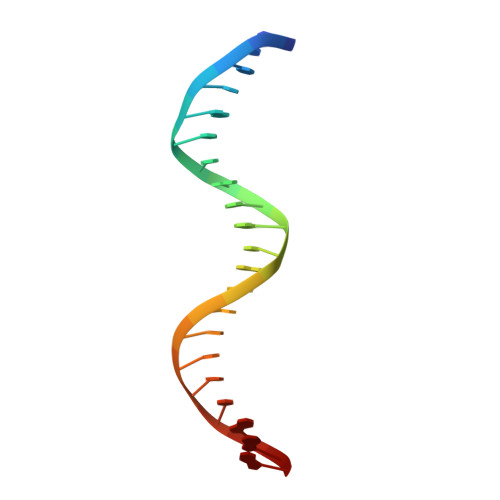
Structure and properties of a re-engineered homeodomain protein-DNA interface.
Simon, M.D., Feldman, M.E., Rauh, D., Maris, A.E., Wemmer, D.E., Shokat, K.M.(2006) ACS Chem Biol 1: 755-760
- PubMed: 17240973
- DOI: https://doi.org/10.1021/cb6003756
- Primary Citation of Related Structures:
2HOS, 2HOT - PubMed Abstract:
The homeodomain (HD)-DNA interface has been conserved over 500 million years of evolution. Despite this conservation, we have successfully re-engineered the engrailed HD to specifically recognize an unnatural nucleotide using a phage display selection. Here we report the synthesis of novel nucleosides and the selection of mutant HDs that bind these nucleotides using phage display. The high-resolution crystal structure of one mutant in complex with modified and unmodified DNA demonstrates that, even with the substantial perturbation to the interface, this selected mutant retains a canonical HD structure. Dissection of the contributions due to each of the selected mutations reveals that the majority of the modification-specific binding is accomplished by a single mutation (I47G) but that the remaining mutations retune the stability of the HD. These results afford a detailed look at a re-engineered protein-DNA interaction and provide insight into the opportunities for re-engineering highly conserved interfaces.