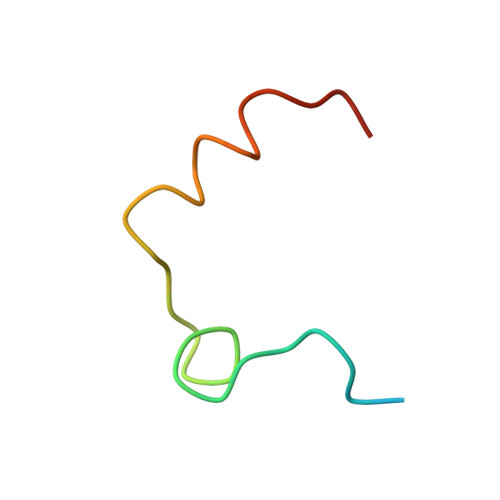
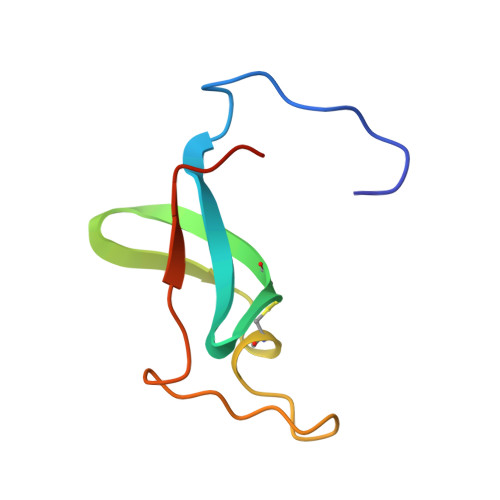
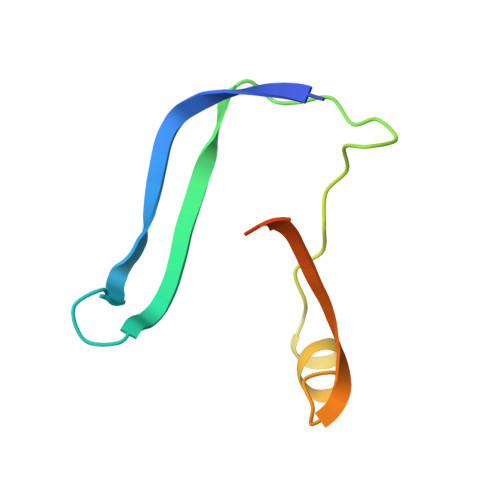
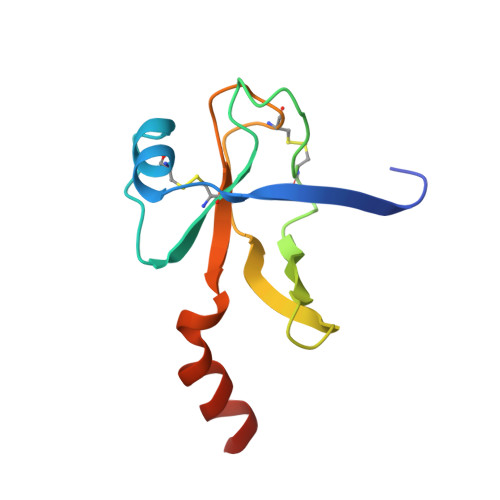
Crystallographic structure of human gamma-thrombin.
Rydel, T.J., Yin, M., Padmanabhan, K.P., Blankenship, D.T., Cardin, A.D., Correa, P.E., Fenton 2nd., J.W., Tulinsky, A.(1994) J Biological Chem 269: 22000-22006
- PubMed: 8071320
- Primary Citation of Related Structures:
2HNT - PubMed Abstract:
In an effort to prepare crystals and determine the structure of alpha-thrombin complexed to a synthetic peptide inhibitor (MDL-28050) of the hirudin 54-65 COOH-terminal region, it was discovered that the crystals were not those of the complex but of gamma-thrombin. Gel electrophoresis studies revealed that autolytic degradation had occurred prior to crystallization. NH2-terminal sequence analysis of these autolytic fragments confirmed the gamma-thrombin product (cleavages at Arg75-Tyr76 and/or Arg77A-Asn78, and Lys149E-Gly150; chymotrypsinogen numbering) with a minor amount of another autolysis product, beta-thrombin (first two cleavages only). The final structure has an R-factor of 0.156 for 7.0-2.5-A data, and includes 186 water molecules. A comparison of gamma-thrombin with the thrombin structure in the alpha-thrombin-hirugen complex revealed that the two structures agreed well (r.m.s. delta = 0.39 A for main chain atoms). These structures possess uninhibited active sites where the disposition of the catalytic triad residues is nearly identical. The electron density in the vicinity of the gamma-thrombin cleavage regions is poor, and only becomes well-defined several residues prior to and after the actual cleavage sites. The extensive disorder evoked by beta-cleavage(s) in the Lys70-Glu80 loop region indicates that this part of the molecule is severely disrupted by autolysis and is the reason exosite functions are dramatically impaired in beta-and gamma-thrombin. Since autolysis did not lead to a major reorganization of the folded structure of alpha-thrombin, the likely structural features of the interaction of thrombin substrate with thrombin enzyme during beta-cleavage have been modeled by docking the exosite region of one molecule at the active site of another.
- Department of Chemistry, Michigan State University, East Lansing 48824-1322.
Organizational Affiliation: