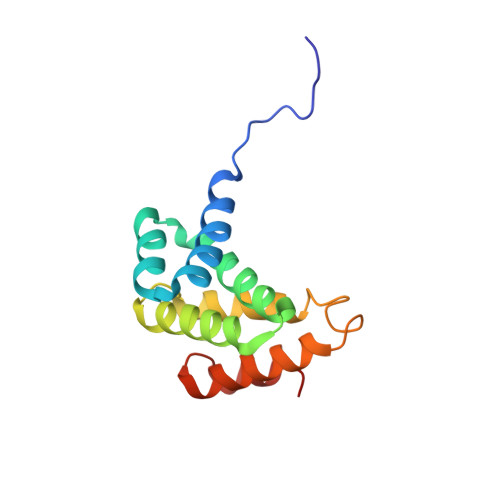
Structure of the C-terminal MA-3 domain of the tumour suppressor protein Pdcd4 and characterization of its interaction with eIF4A
Waters, L.C., Veverka, V., Bohm, M., Schmedt, T., Choong, P.T., Muskett, F.W., Klempnauer, K.H., Carr, M.D.(2007) Oncogene 26: 4941-4950
- PubMed: 17310995
- DOI: https://doi.org/10.1038/sj.onc.1210305
- Primary Citation of Related Structures:
2HM8 - PubMed Abstract:
Programmed cell death protein 4 (Pdcd4) is a novel tumour suppressor protein, which is involved in the control of eukaryotic transcription and translation. The regulation of translation involves specific interactions with eukaryotic initiation factor (eIF)4A and eIF4G, which are mediated via the two tandem MA-3 domains. We have determined the structure of the C-terminal MA-3 domain of Pdcd4 (Pdcd4 MA-3(C)), characterized its interaction with eIF4A and compared the features of nuclear magnetic resonance (NMR) spectra obtained from the single domain and tandem MA-3 region. Pdcd4 MA-3(C) is composed of three layers of helix-turn-helix hairpins capped by a single helix and shows close structural homology to the atypical HEAT repeats found in many eIFs. The sequence conservation and NMR data strongly suggest that the tandem MA-3 region is composed of two equivalent domains connected by a somewhat flexible linker. Pdcd4 MA-3(C) was found to interact with the N-terminal domain of eIF4A through a conserved surface region encompassing the loop connecting alpha5 and alpha6 and the turn linking alpha3 and alpha4. This site is strongly conserved in other MA-3 domains known to interact with eIF4A, including the preceding domain of Pdcd4, suggesting a common mode of binding.
- Department of Biochemistry, University of Leicester, Leicester, UK.
Organizational Affiliation: