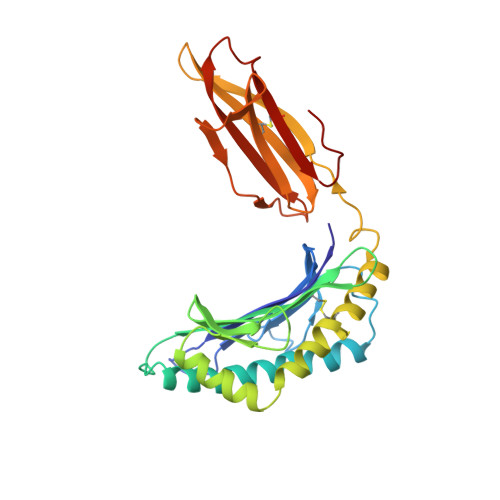
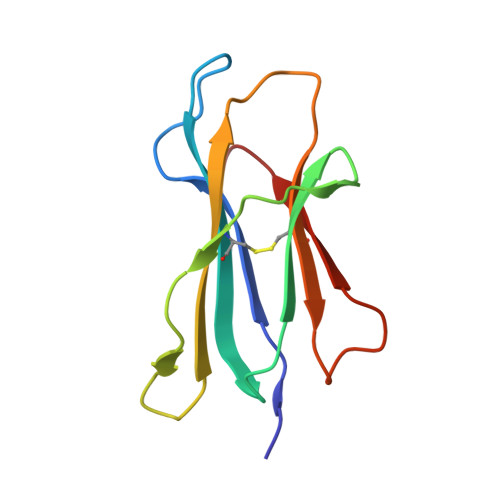
Specificity pockets for the side chains of peptide antigens in HLA-Aw68.
Garrett, T.P., Saper, M.A., Bjorkman, P.J., Strominger, J.L., Wiley, D.C.(1989) Nature 342: 692-696
- PubMed: 2594067
- DOI: https://doi.org/10.1038/342692a0
- Primary Citation of Related Structures:
2HLA - PubMed Abstract:
We have determined the structure of a second human histocompatibility glycoprotein, HLA-Aw68, by X-ray crystallography and refined it to a resolution of 2.6 A. Overall, the structure is extremely similar to that of HLA-A2 (refs 1, 2; and M.A.S. et al., manuscript in preparation), although the 11 amino-acid substitutions at polymorphic residues in the antigen-binding cleft alter the detailed shape and electrostatic charge of that site. A prominent negatively charged pocket within the cleft extends underneath the alpha-helix of the alpha 1-domain, providing a potential subsite for recognizing a positively charged side chain or peptide N terminus. Uninterpreted electron density, presumably representing an unknown 'antigen(s)', which seems to be different from that seen in the HLA-A2 structure, occupies the cleft and extends into the negatively charged pocket in HLA-Aw68. The structures of HLA-Aw68 and HLA-A2 demonstrate how polymorphism creates and alters subsites (pockets) positioned to bind peptide side chains, thereby suggesting the structural basis for allelic specificity in foreign antigen binding.
- Department of Biochemistry and Molecular Biology, Harvard University, Cambridge, Massachusetts 02138.
Organizational Affiliation: