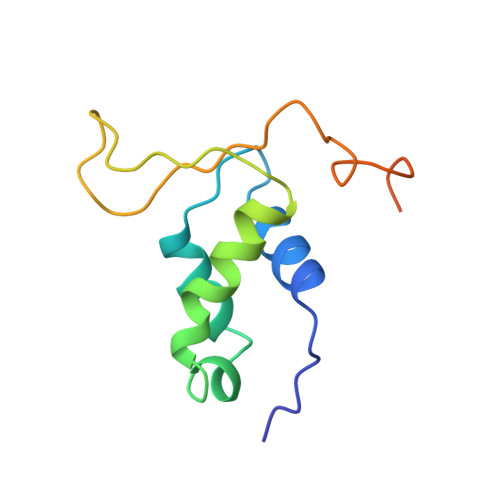
Structural changes in the region directly adjacent to the DNA-binding helix highlight a possible mechanism to explain the observed changes in the sequence-specific binding of winged helix proteins.
Marsden, I., Jin, C., Liao, X.(1998) J Mol Biology 278: 293-299
- PubMed: 9571051
- DOI: https://doi.org/10.1006/jmbi.1998.1703
- Primary Citation of Related Structures:
2HFH - PubMed Abstract:
The hepatocyte nuclear factor 3 (HNF-3)/fork head (fkh) family contains a large number of transcription factors and folds into a winged helix motif. Despite having almost invariable amino acid sequences in their principal DNA-binding helices, HNF-3/fkh proteins show a wide diversity of sequence-specific binding. Previous studies of chimeric HNF-3/fkh proteins demonstrated that the binding specificity is primarily influenced by a region directly adjacent to the binding helix. We report our findings of an NMR structural study performed on an HNF-3/fkh family member (Genesis, formerly HFH-2) and compare it to that of another family member (HNF-3gamma) complexed to DNA and determined by X-ray crystallography. It is found that in comparison to HNF-3gamma, Genesis contains an extra small helix directly prior to the N terminus of the primary DNA contact helix. Due to the insertion of this helix, a shorter and slightly re-positioned primary DNA contact helix is observed, which we believe leads to the DNA-binding specificity differences among family members.
- College of Medicine, University of Illinois at Chicago, 1819 West Polk Street, Chicago, IL 60612, USA.
Organizational Affiliation: