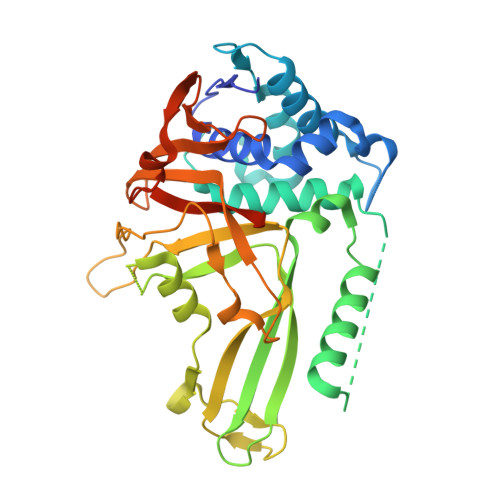
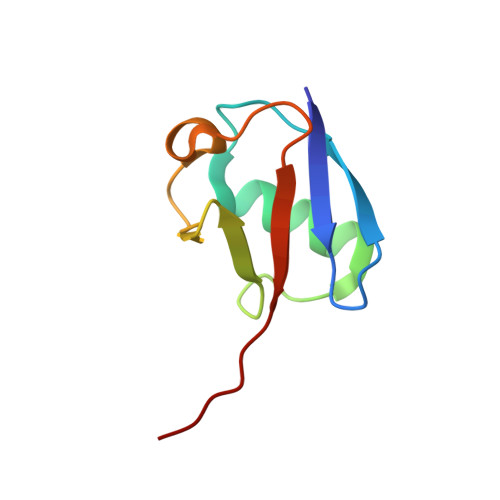
Structural Basis of Ubiquitin Recognition by the Deubiquitinating Protease USP2.
Renatus, M., Parrado, S.G., D'Arcy, A., Eidhoff, U., Gerhartz, B., Hassiepen, U., Pierrat, B., Riedl, R., Vinzenz, D., Worpenberg, S., Kroemer, M.(2006) Structure 14: 1293-1302
- PubMed: 16905103
- DOI: https://doi.org/10.1016/j.str.2006.06.012
- Primary Citation of Related Structures:
2HD5 - PubMed Abstract:
Deubiquitinating proteases reverse protein ubiquitination and rescue their target proteins from destruction by the proteasome. USP2, a cysteine protease and a member of the ubiquitin specific protease family, is overexpressed in prostate cancer and stabilizes fatty acid synthase, which has been associated with the malignancy of some aggressive prostate cancers. Here, we report the structure of the human USP2 catalytic domain in complex with ubiquitin. Ubiquitin uses two major sites for the interaction with the protease. Both sites are required simultaneously, as shown by USP2 inhibition assays with peptides and ubiquitin mutants. In addition, a layer of ordered water molecules mediates key interactions between ubiquitin and USP2. As several of those molecules are found at identical positions in the previously solved USP7/ubiquitin-aldehyde complex structure, we suggest a general mechanism of water-mediated ubiquitin recognition by USPs.
- Protease Platform, Novartis Institutes for BioMedical Research, 4002 Basel, Switzerland. martin.renatus@novartis.com
Organizational Affiliation: