A Common Allosteric Site and Mechanism in Caspases
Scheer, J.M., Romanowski, M.J., Wells, J.A.(2006) Proc Natl Acad Sci U S A 103: 7595-7600
- PubMed: 16682620
- DOI: https://doi.org/10.1073/pnas.0602571103
- Primary Citation of Related Structures:
2FQQ, 2H48, 2HBQ, 2HBR, 2HBY, 2HBZ - PubMed Abstract:
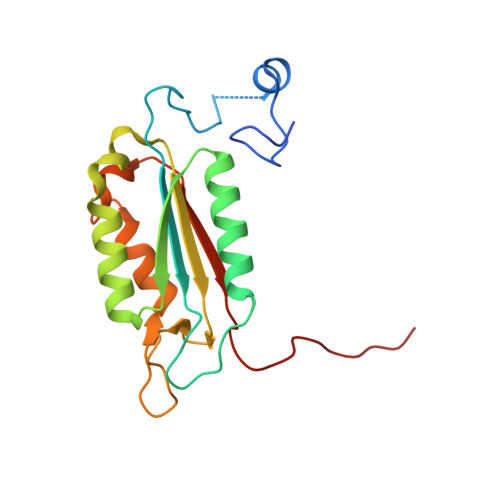
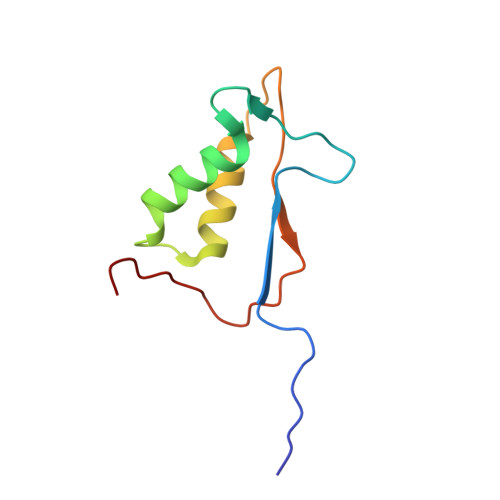
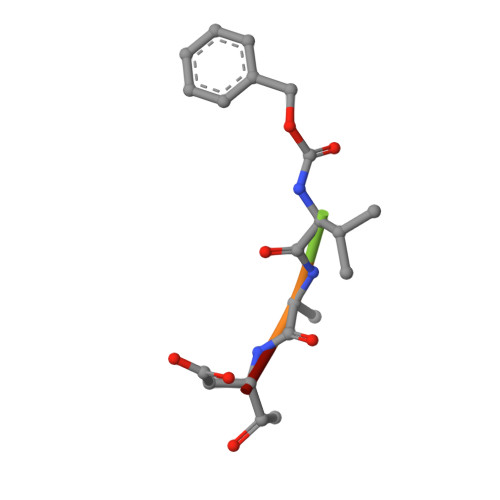
We present a common allosteric mechanism for control of inflammatory and apoptotic caspases. Highly specific thiol-containing inhibitors of the human inflammatory caspase-1 were identified by using disulfide trapping, a method for site-directed small-molecule discovery. These compounds became trapped by forming a disulfide bond with a cysteine residue in the cavity at the dimer interface approximately 15 A away from the active site. Mutational and structural analysis uncovered a linear circuit of functional residues that runs from one active site through the allosteric cavity and into the second active site. Kinetic analysis revealed robust positive cooperativity not seen in other endopeptidases. Recently, disulfide trapping identified a similar small-molecule site and allosteric transition in the apoptotic caspase-7 that shares only a 23% sequence identity with caspase-1. Together, these studies show a general small-molecule-binding site for functionally reversing the zymogen activation of caspases and suggest a common regulatory site for the allosteric control of inflammation and apoptosis.
- Department of Pharmaceutical Chemistry, University of California, 1700 4th Street, San Francisco, CA 94143, USA.
Organizational Affiliation: