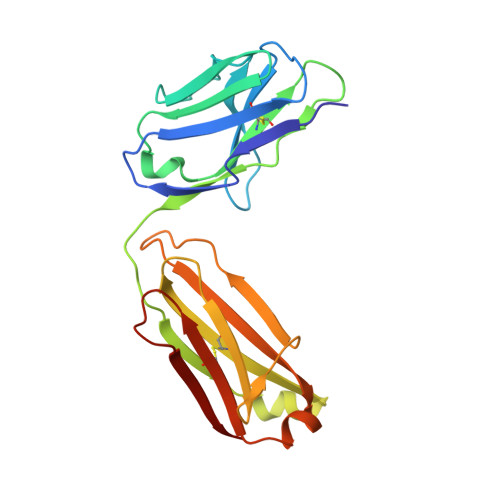
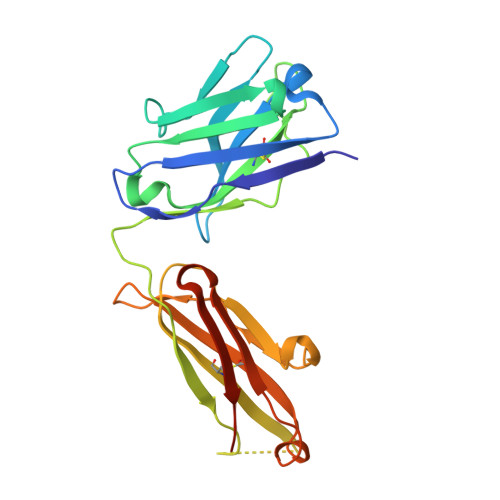
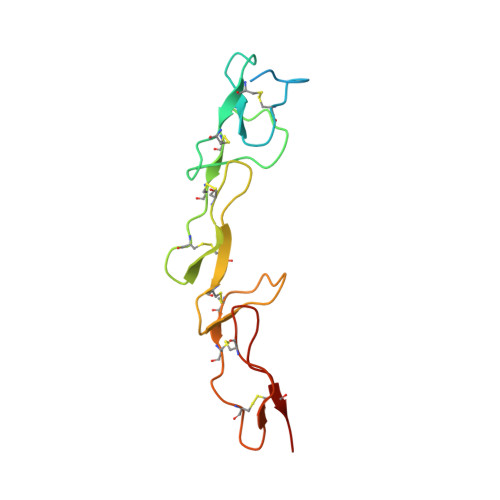
Activation of the Proapoptotic Death Receptor DR5 by Oligomeric Peptide and Antibody Agonists.
Li, B., Russell, S.J., Compaan, D.M., Totpal, K., Marsters, S.A., Ashkenazi, A., Cochran, A.G., Hymowitz, S.G., Sidhu, S.S.(2006) J Mol Biology 361: 522-536
- PubMed: 16859704
- DOI: https://doi.org/10.1016/j.jmb.2006.06.042
- Primary Citation of Related Structures:
2H9G - PubMed Abstract:
The cell-extrinsic apoptotic pathway triggers programmed cell death in response to certain ligands that bind to cell-surface death receptors. Apoptosis is essential for normal development and homeostasis in metazoans, and furthermore, selective activation of the cell-extrinsic pathway in tumor cells holds considerable promise for cancer therapy. We used phage display to identify peptides and synthetic antibodies that specifically bind to the human proapoptotic death receptor DR5. Despite great differences in overall size and structure, the DR5-binding peptides and antibodies shared a tripeptide motif, which was conserved within a disulfide-constrained loop of the peptides and the third complementarity determining region of the antibody heavy chains. The X-ray crystal structure of an antibody in complex with DR5 revealed that the tripeptide motif is buried at the core of the interface, confirming its central role in antigen recognition. We found that certain peptides and antibodies exhibited potent proapoptotic activity against DR5-expressing SK-MES-1 lung carcinoma cells. These phage-derived ligands may be useful for elucidating DR5 activation at the molecular level and for creating synthetic agonists of proapoptotic death receptors.
- Department of Protein Engineering, Genentech Inc., 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: