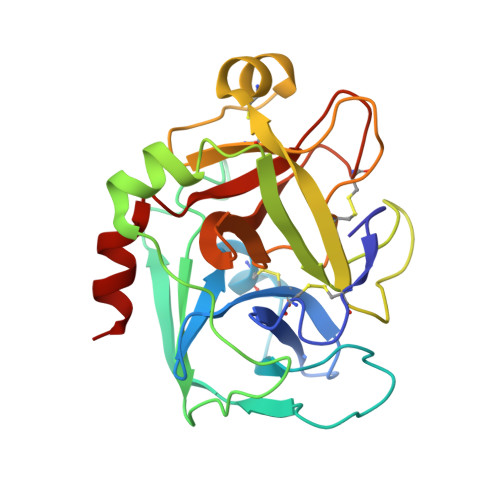
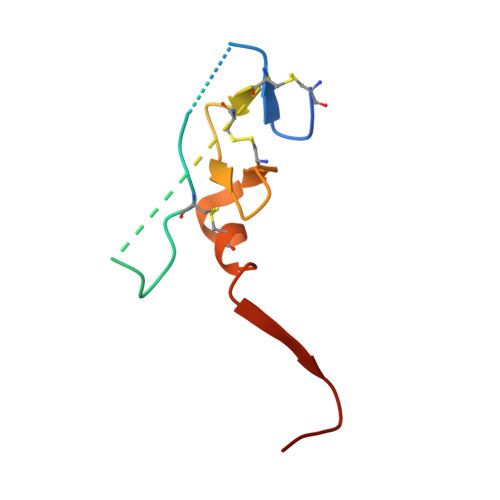
Intermolecular Interactions and Characterization of the Novel Factor Xa Exosite Involved in Macromolecular Recognition and Inhibition: Crystal Structure of Human Gla-domainless Factor Xa Complexed with the Anticoagulant Protein NAPc2 from the Hematophagous Nematode Ancylostoma caninum.
Murakami, M.T., Rios-Steiner, J., Weaver, S.E., Tulinsky, A., Geiger, J.H., Arni, R.K.(2007) J Mol Biology 366: 602-610
- PubMed: 17173931
- DOI: https://doi.org/10.1016/j.jmb.2006.11.040
- Primary Citation of Related Structures:
2H9E - PubMed Abstract:
NAPc2, an anticoagulant protein from the hematophagous nematode Ancylostoma caninum evaluated in phase-II/IIa clinical trials, inhibits the extrinsic blood coagulation pathway by a two step mechanism, initially interacting with the hitherto uncharacterized factor Xa exosite involved in macromolecular recognition and subsequently inhibiting factor VIIa (K(i)=8.4 pM) of the factor VIIa/tissue factor complex. NAPc2 is highly flexible, becoming partially ordered and undergoing significant structural changes in the C terminus upon binding to the factor Xa exosite. In the crystal structure of the ternary factor Xa/NAPc2/selectide complex, the binding interface consists of an intermolecular antiparallel beta-sheet formed by the segment of the polypeptide chain consisting of residues 74-80 of NAPc2 with the residues 86-93 of factor Xa that is additional maintained by contacts between the short helical segment (residues 67-73) and a turn (residues 26-29) of NAPc2 with the short C-terminal helix of factor Xa (residues 233-243). This exosite is physiologically highly relevant for the recognition and inhibition of factor X/Xa by macromolecular substrates and provides a structural motif for the development of a new class of inhibitors for the treatment of deep vein thrombosis and angioplasty.
- Department of Physics, IBILCE/UNESP, São José do Rio Preto, SP 15054-000, Brazil.
Organizational Affiliation: