Differences in Binding Specificity for the Homologous gamma- and beta-Chain "Holes" on Fibrinogen: Exclusive Binding of Ala-His-Arg-Pro-amide by the beta-Chain Hole.
Doolittle, R.F., Chen, A., Pandi, L.(2006) Biochemistry 45: 13962-13969
- PubMed: 17115691
- DOI: https://doi.org/10.1021/bi061219e
- Primary Citation of Related Structures:
2H43 - PubMed Abstract:
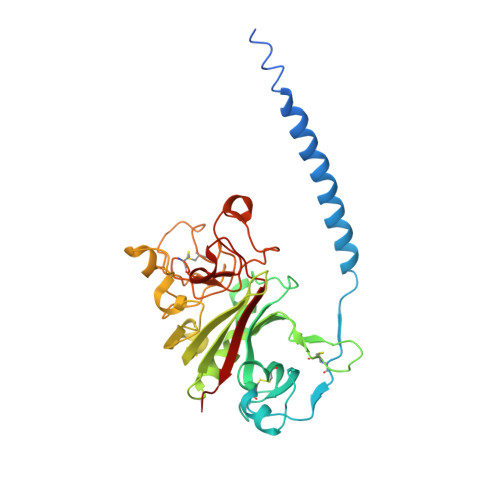
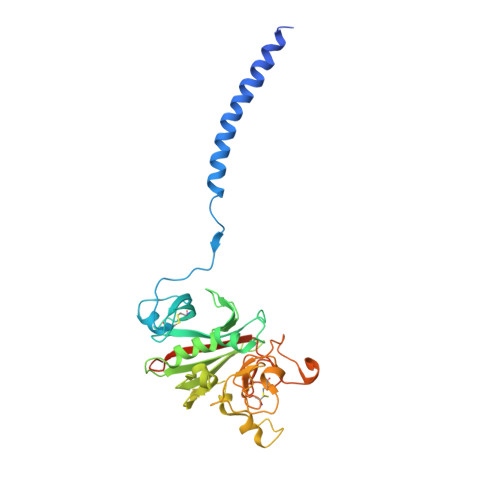
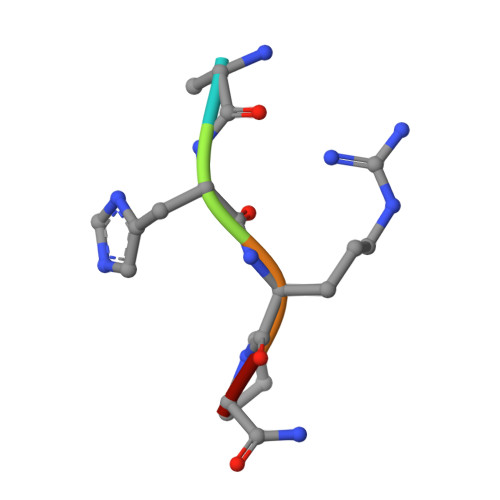
The beta-chain amino-terminal sequences of all known mammalian fibrins begin with the sequence Gly-His-Arg-Pro- (GHRP-), but the homologous sequence in chicken fibrin begins with the sequence Ala-His-Arg-Pro- (AHRP-). Nonetheless, chicken fibrinogen binds the synthetic peptide GHRPam, and a previously reported crystal structure has revealed that the binding is in exact conformance with that observed for the human GHRPam-fragment D complex. We now report that human fibrinogen, which is known not to bind APRP, binds the synthetic peptide AHRPam. Moreover, a crystal structure of AHRPam complexed with fragment D from human fibrinogen shows that AHRPam binds exclusively to the beta-chain hole and, unlike GHRPam, not at all to the homologous gamma-chain hole. The difference can be attributed to the methyl group of the alanine residue clashing with a critical carboxyl group in the gammaC hole but being accommodated in the roomier betaC hole where the equivalent carboxyl is situated more flexibly.
- Department of Chemistry and Biochemistry and Division of Biology, University of California at San Diego, La Jolla, California 92093-0314, USA. rdoolittle@ucsd.edu
Organizational Affiliation: