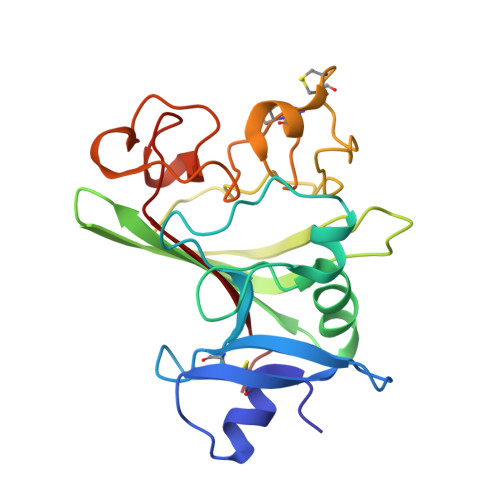
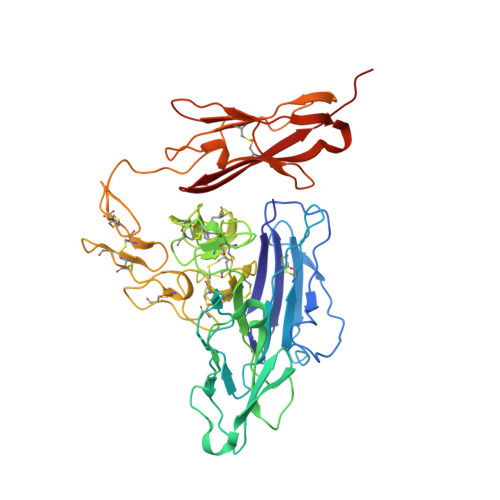
Crystal structures of the Tie2 receptor ectodomain and the angiopoietin-2-Tie2 complex.
Barton, W.A., Tzvetkova-Robev, D., Miranda, E.P., Kolev, M.V., Rajashankar, K.R., Himanen, J.P., Nikolov, D.B.(2006) Nat Struct Mol Biol 13: 524-532
- PubMed: 16732286
- DOI: https://doi.org/10.1038/nsmb1101
- Primary Citation of Related Structures:
2GY5, 2GY7 - PubMed Abstract:
The Tie receptor tyrosine kinases and their angiopoietin (Ang) ligands play central roles in developmental and tumor-induced angiogenesis. Here we present the crystal structures of the Tie2 ligand-binding region alone and in complex with Ang2. In contrast to prediction, Tie2 contains not two but three immunoglobulin (Ig) domains, which fold together with the three epidermal growth factor domains into a compact, arrowhead-shaped structure. Ang2 binds at the tip of the arrowhead utilizing a lock-and-key mode of ligand recognition-unique for a receptor kinase-where two complementary surfaces interact with each other with no domain rearrangements and little conformational change in either molecule. Ang2-Tie2 recognition is similar to antibody-protein antigen recognition, including the location of the ligand-binding site within the Ig fold. Analysis of the structures and structure-based mutagenesis provide insight into the mechanism of receptor activation and support the hypothesis that all angiopoietins interact with Tie2 in a structurally similar manner.
- Structural Biology Program, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, New York 10021, USA.
Organizational Affiliation: