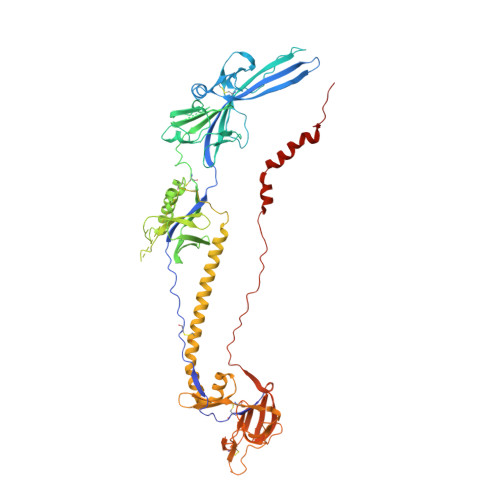
Crystal structure of glycoprotein B from herpes simplex virus 1.
Heldwein, E.E., Lou, H., Bender, F.C., Cohen, G.H., Eisenberg, R.J., Harrison, S.C.(2006) Science 313: 217-220
- PubMed: 16840698
- DOI: https://doi.org/10.1126/science.1126548
- Primary Citation of Related Structures:
2GUM - PubMed Abstract:
Glycoprotein B (gB) is the most conserved component of the complex cell-entry machinery of herpes viruses. A crystal structure of the gB ectodomain from herpes simplex virus type 1 reveals a multidomain trimer with unexpected homology to glycoprotein G from vesicular stomatitis virus (VSV G). An alpha-helical coiled-coil core relates gB to class I viral membrane fusion glycoproteins; two extended beta hairpins with hydrophobic tips, homologous to fusion peptides in VSV G, relate gB to class II fusion proteins. Members of both classes accomplish fusion through a large-scale conformational change, triggered by a signal from a receptor-binding component. The domain connectivity within a gB monomer would permit such a rearrangement, including long-range translocations linked to viral and cellular membranes.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 320 Longwood Avenue, Boston, MA 02115, USA. heldwein@crystal.harvard.edu
Organizational Affiliation: