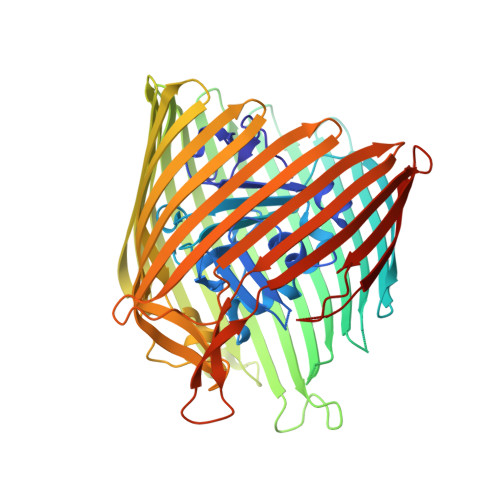
In Meso Structure of the Cobalamin Transporter, BtuB, at 1.95 A Resolution.
Cherezov, V., Yamashita, E., Liu, W., Zhalnina, M., Cramer, W.A., Caffrey, M.(2006) J Mol Biology 364: 716-734
- PubMed: 17028020
- DOI: https://doi.org/10.1016/j.jmb.2006.09.022
- Primary Citation of Related Structures:
2GUF - PubMed Abstract:
Crystals of the apo form of the vitamin B12 and colicin receptor, BtuB, that diffract to 1.95 A have been grown by the membrane-based in meso technique. The structure of the protein differs in several details from that of its counterpart grown by the more traditional, detergent-based (in surfo) method. Some of these differences include (i) the five N-terminal residues are resolved in meso, (ii) residues 57-62 in the hatch domain and residues 574-581 in loop 21-22 are disordered in meso and are ordered in surfo, (iii) residues 278-287 in loop 7-8 are resolved in meso, (iv) residues 324-331 in loop 9-10, 396-411 in loop 13-14, 442-458 in loop 15-16 and 526-541 in loop 19-20 have large differences in position between the two crystal forms, as have residues 86-96 in the hatch domain, and (v) the conformation of residues 6 and 7 in the Ton box (considered critical to signal transduction and substrate transport) are entirely different in the two structures. Importantly, the in meso orientation of residues 6 and 7 is similar to that of the vitamin B12-charged state. These data suggest that the "substrate-induced" 180 degrees -rotation of residues 6 and 7 reported in the literature may not be a unique signalling event. The extent to which these findings agree with structural, dynamic and functional insights gleaned from site-directed spin labelling and electron paramagnetic resonance measurements is evaluated. Packing in in meso grown crystals is dense and layered, consistent with the current model for crystallogenesis of membrane proteins in lipidic mesophases. Layered packing has been used to locate the transmembrane hydrophobic surface of the protein. Generally, this is consistent with tryptophan, tyrosine, lipid and CalphaB-factor distributions in the protein, and with predictions based on transfer free energy calculations.
- Department of Chemistry, The Ohio State University, Columbus, OH 43210, USA.
Organizational Affiliation: