Lumbricus erythrocruorin at 3.5 a resolution: architecture of a megadalton respiratory complex.
Royer Jr., W.E., Sharma, H., Strand, K., Knapp, J.E., Bhyravbhatla, B.(2006) Structure 14: 1167-1177
- PubMed: 16843898
- DOI: https://doi.org/10.1016/j.str.2006.05.011
- Primary Citation of Related Structures:
2GTL - PubMed Abstract:
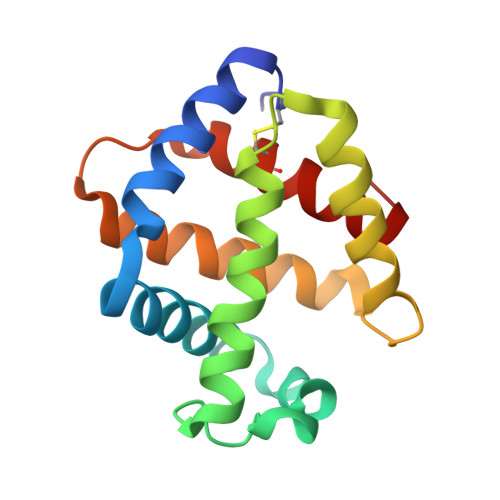
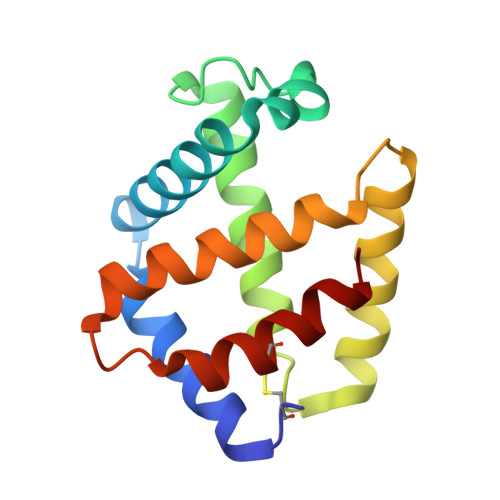
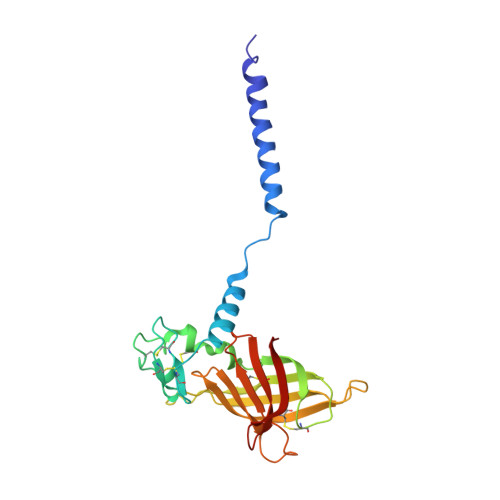
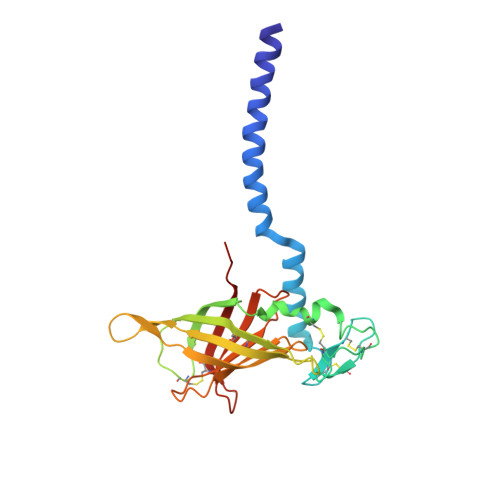
Annelid erythrocruorins are highly cooperative extracellular respiratory proteins with molecular masses on the order of 3.6 million Daltons. We report here the 3.5 A crystal structure of erythrocruorin from the earthworm Lumbricus terrestris. This structure reveals details of symmetrical and quasi-symmetrical interactions that dictate the self-limited assembly of 144 hemoglobin and 36 linker subunits. The linker subunits assemble into a core complex with D(6) symmetry onto which 12 hemoglobin dodecamers bind to form the entire complex. Although the three unique linker subunits share structural similarity, their interactions with each other and the hemoglobin subunits display striking diversity. The observed diversity includes design features that have been incorporated into the linker subunits and may be critical for efficient assembly of large quantities of this complex respiratory protein.
- Department of Biochemistry and Molecular Pharmacology, University of Massachusetts Medical School, Worcester, MA 01655, USA. william.royer@umassmed.edu
Organizational Affiliation: