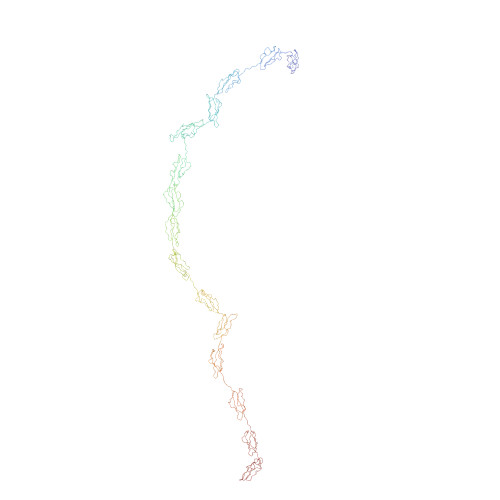The 15 SCR Flexible Extracellular Domains of Human Complement Receptor Type 2 can Mediate Multiple Ligand and Antigen Interactions.
Gilbert, H.E., Asokan, R., Holers, V.M., Perkins, S.J.(2006) J Mol Biology 362: 1132-1147
- PubMed: 16950392
- DOI: https://doi.org/10.1016/j.jmb.2006.08.012
- Primary Citation of Related Structures:
2GSX - PubMed Abstract:
Complement receptor type 2 (CR2, CD21) is a cell surface protein that links the innate and adaptive immune response during the activation of B cells. The extracellular portion of CR2 comprises 15 or 16 short complement regulator (SCR) domains, for which the overall arrangement in solution is unknown. This was determined by constrained scattering and ultracentrifugation modelling. The radius of gyration of CR2 SCR 1-15 was determined to be 11.5 nm by both X-ray and neutron scattering, and that of its cross-section was 1.8 nm. The distance distribution function P(r) showed that the overall length of CR2 SCR 1-15 was 38 nm. Sedimentation equilibrium curve fits gave a mean molecular weight of 135,000 (+/- 13,000) Da, in agreement with a fully glycosylated structure. Velocity experiments using the g*(s) derivative method gave a sedimentation coefficient of 4.2 (+/- 0.1) S. In order to construct a model of CR2 SCR 1-15 for constrained fitting, homology models for the 15 SCR domains were combined with randomised linker peptides generated by molecular dynamics simulations. Using an automated procedure, the analysis of 15,000 possible CR2 SCR 1-15 models showed that only those models in which the 15 SCR domains were flexible but partially folded back accounted for the scattering and sedimentation data. The best-fit CR2 models provided a visual explanation for the versatile interaction of CR2 with four ligands C3d, CD23, gp350 and IFN-alpha. The flexible location of CR2 SCR 1-2 is likely to facilitate interactions of C3d-antigen complexes with the B cell receptor.
- Department of Biochemistry and Molecular Biology, Darwin Building, University College London, Gower Street, London, WC1E 6BT, UK.
Organizational Affiliation: