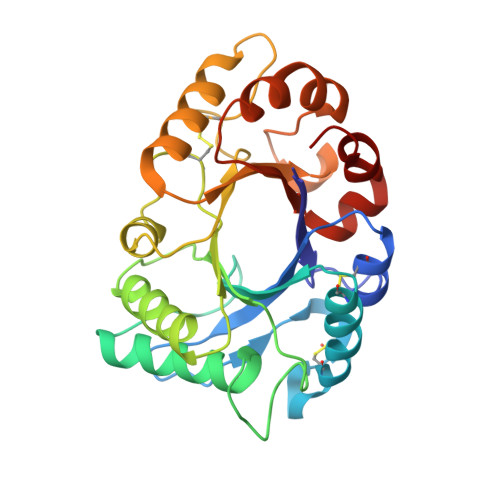
cDNA cloning and 1.75 A crystal structure determination of PPL2, an endochitinase and N-acetylglucosamine-binding hemagglutinin from Parkia platycephala seeds
Cavada, B.S., Moreno, F.B., da Rocha, B.A., de Azevedo Jr., W.F., Castellon, R.E.R., Goersch, G.V., Nagano, C.S., de Souza, E.P., Nascimento, K.S., Radis-Baptista, G., Delatorre, P., Leroy, Y., Toyama, M.H., Pinto, V.P., Sampaio, A.H., Barettino, D., Debray, H., Calvete, J.J., Sanz, L.(2006) FEBS J 273: 3962-3974
- PubMed: 16934035
- DOI: https://doi.org/10.1111/j.1742-4658.2006.05400.x
- Primary Citation of Related Structures:
2GSJ - PubMed Abstract:
Parkia platycephala lectin 2 was purified from Parkia platycephala (Leguminosae, Mimosoideae) seeds by affinity chromatography and RP-HPLC. Equilibrium sedimentation and MS showed that Parkia platycephala lectin 2 is a nonglycosylated monomeric protein of molecular mass 29 407+/-15 Da, which contains six cysteine residues engaged in the formation of three intramolecular disulfide bonds. Parkia platycephala lectin 2 agglutinated rabbit erythrocytes, and this activity was specifically inhibited by N-acetylglucosamine. In addition, Parkia platycephala lectin 2 hydrolyzed beta(1-4) glycosidic bonds linking 2-acetoamido-2-deoxy-beta-D-glucopyranose units in chitin. The full-length amino acid sequence of Parkia platycephala lectin 2, determined by N-terminal sequencing and cDNA cloning, and its three-dimensional structure, established by X-ray crystallography at 1.75 A resolution, showed that Parkia platycephala lectin 2 is homologous to endochitinases of the glycosyl hydrolase family 18, which share the (betaalpha)8 barrel topology harboring the catalytic residues Asp125, Glu127, and Tyr182.
- BioMol-Laboratory, Departamento de Bioquímica e Biologia Molecular, Universidade Federal do Ceará, Fortaleza, Ceará, Brazil. bscavada@ufc.br
Organizational Affiliation: