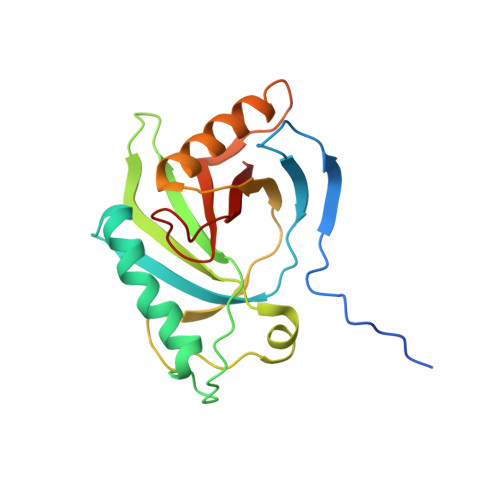
The First Structure from the SOUL/HBP Family of Heme-binding Proteins, Murine P22HBP.
Dias, J.S., Macedo, A.L., Ferreira, G.C., Peterson, F.C., Volkman, B.F., Goodfellow, B.J.(2006) J Biological Chem 281: 31553-31561
- PubMed: 16905545
- DOI: https://doi.org/10.1074/jbc.M605988200
- Primary Citation of Related Structures:
2GOV - PubMed Abstract:
Murine p22HBP, a 22-kDa monomer originally identified as a cytosolic heme-binding protein ubiquitously expressed in various tissues, has 27% sequence identity to murine SOUL, a heme-binding hexamer specifically expressed in the retina. In contrast to murine SOUL, which binds one heme per subunit via coordination of the Fe(III)-heme to a histidine, murine p22HBP binds one heme molecule per subunit with no specific axial ligand coordination of the Fe(III)-heme. Using intrinsic protein fluorescence quenching, the values for the dissociation constants of p22HBP for hemin and protoporphyrin-IX were determined to be in the low nanomolar range. The three-dimensional structure of murine p22HBP, the first for a protein from the SOUL/HBP family, was determined by NMR methods to consist of a 9-stranded distorted beta-barrel flanked by two long alpha-helices. Although homologous domains have been found in three bacterial proteins, two of which are transcription factors, the fold determined for p22HBP corresponds to a novel alpha plus beta fold in a eukaryotic protein. Chemical shift mapping localized the tetrapyrrole binding site to a hydrophobic cleft formed by residues from helix alphaA and an extended loop. In an attempt to assess the structural basis for tetrapyrrole binding in the SOUL/HBP family, models for the p22HBP-protoporphyrin-IX complex and the SOUL protein were generated by manual docking and automated methods.
- Rede de Química e Tecnologia, Centro de Química Fina e Biotecnologia, Departamento de Química, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa, 2829-516 Caparica, Portugal.
Organizational Affiliation: