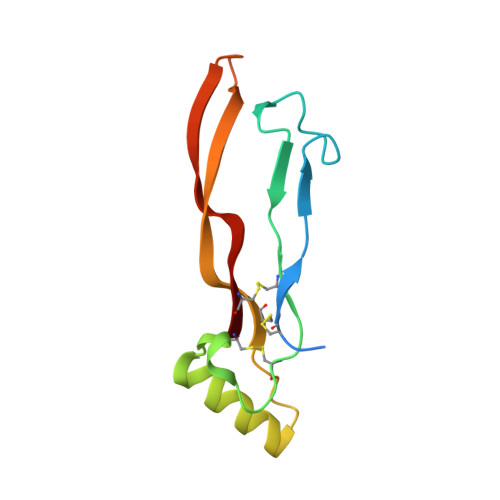
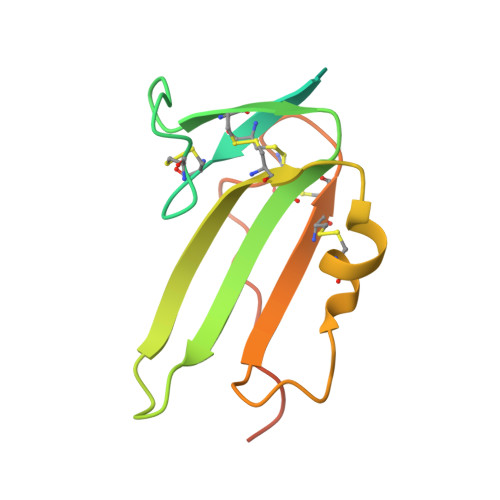
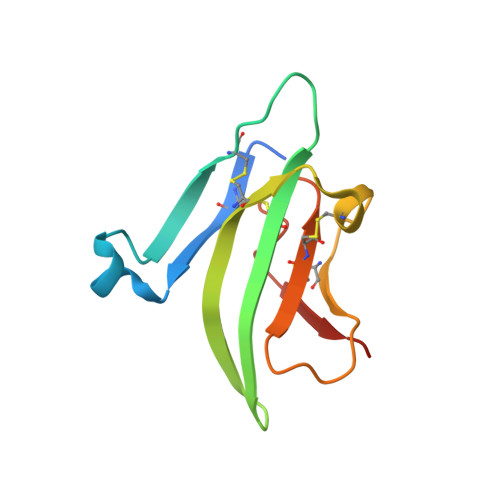
Structure of the ternary signaling complex of a TGF-beta superfamily member.
Allendorph, G.P., Vale, W.W., Choe, S.(2006) Proc Natl Acad Sci U S A 103: 7643-7648
- PubMed: 16672363
- DOI: https://doi.org/10.1073/pnas.0602558103
- Primary Citation of Related Structures:
2GOO - PubMed Abstract:
The crystal structure of the complete signaling complex formed between bone morphogenetic protein 2 (BMP-2) and the extracellular domains (ECDs) of its type I receptor [bone morphogenetic protein receptor type Ia (BMPR-Ia)-ECD] and its type II receptor [activin receptor type II (ActRII)-ECD] shows two fundamental structural constraints for receptor assembly. First, the homodimeric BMP-2 ligand assembles two pairs of each receptor symmetrically, where each of the receptor ECDs does not make physical contact. Therefore, conformational communication between receptor ECDs, if any, should be propagated through the central ligand. Second, the type I and II receptor interfaces of the complex, when compared with those of binary complexes such as BMP-2/BMPR Ia-ECD, BMP-7/ActRII-ECD, and activin/ActRIIb-ECD, respectively, show there are common sets of positions repeatedly used by both ligands and receptors. Therefore, specificity-determining amino acid differences at the receptor interfaces should also account for the disparity in affinity of individual receptors for different ligand subunits. We find that a specific mutation to BMP-2 increases its affinity to ActRII-ECD by 5-fold. These results together establish that the specific signaling output is largely determined by two variables, the ligand-receptor pair identity and the mode of cooperative assembly of relevant receptors governed by the ligand flexibility in a membrane-restricted manner.
- Structural Biology Laboratory, The Salk Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: