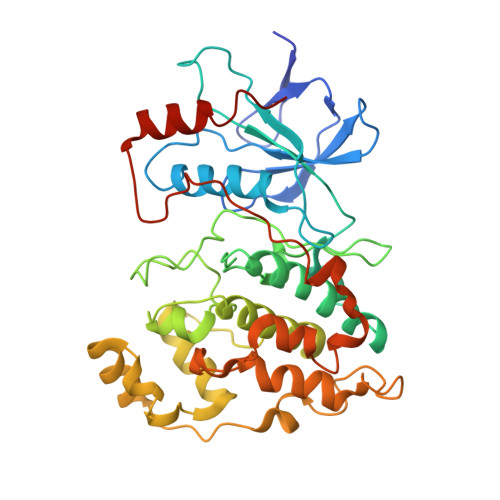
Aminopyridine-Based c-Jun N-Terminal Kinase Inhibitors with Cellular Activity and Minimal Cross-Kinase Activity.
Szczepankiewicz, B.G., Kosogof, C., Nelson, L.T., Liu, G., Liu, B., Zhao, H., Serby, M.D., Xin, Z., Liu, M., Gum, R.J., Haasch, D.L., Wang, S., Clampit, J.E., Johnson, E.F., Lubben, T.H., Stashko, M.A., Olejniczak, E.T., Sun, C., Dorwin, S.A., Haskins, K., Abad-Zapatero, C., Fry, E.H., Hutchins, C.W., Sham, H.L., Rondinone, C.M., Trevillyan, J.M.(2006) J Med Chem 49: 3563-3580
- PubMed: 16759099
- DOI: https://doi.org/10.1021/jm060199b
- Primary Citation of Related Structures:
2GMX - PubMed Abstract:
The c-Jun N-terminal kinases (JNK-1, -2, and -3) are members of the mitogen activated protein (MAP) kinase family of enzymes. They are activated in response to certain cytokines, as well as by cellular stresses including chemotoxins, peroxides, and irradiation. They have been implicated in the pathology of a variety of different diseases with an inflammatory component including asthma, stroke, Alzheimer's disease, and type 2 diabetes mellitus. In this work, high-throughput screening identified a JNK inhibitor with an excellent kinase selectivity profile. Using X-ray crystallography and biochemical screening to guide our lead optimization, we prepared compounds with inhibitory potencies in the low-double-digit nanomolar range, activity in whole cells, and pharmacokinetics suitable for in vivo use. The new compounds were over 1,000-fold selective for JNK-1 and -2 over other MAP kinases including ERK2, p38alpha, and p38delta and showed little inhibitory activity against a panel of 74 kinases.
- Metabolic Disease Research, Global Pharmaceutical Research and Discovery Organization, Abbott Laboratories, 100 Abbott Park Road, Abbott Park, Illinois 60064-6098, USA. bruce.szczepankiewicz@abbott.com
Organizational Affiliation: