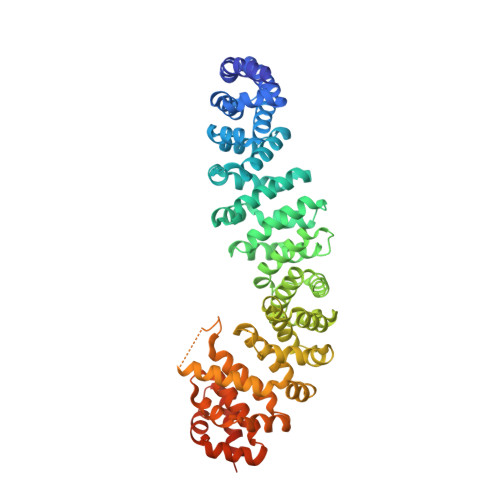
Crystal Structure of a beta-Catenin/BCL9/Tcf4 Complex.
Sampietro, J., Dahlberg, C.L., Cho, U.S., Hinds, T.R., Kimelman, D., Xu, W.(2006) Mol Cell 24: 293-300
- PubMed: 17052462
- DOI: https://doi.org/10.1016/j.molcel.2006.09.001
- Primary Citation of Related Structures:
2GL7 - PubMed Abstract:
The canonical Wnt pathway plays critical roles in embryonic development, stem cell growth, and tumorigenesis. Stimulation of the Wnt pathway leads to the association of beta-catenin with Tcf and BCL9 in the nucleus, resulting in the transactivation of Wnt target genes. We have determined the crystal structure of a beta-catenin/BCL9/Tcf-4 triple complex at 2.6 A resolution. Our studies reveal that the beta-catenin binding site of BCL9 is distinct from that of most other beta-catenin partners and forms a good target for developing drugs that block canonical Wnt/beta-catenin signaling. The BCL9 beta-catenin binding domain (CBD) forms an alpha helix that binds to the first armadillo repeat of beta-catenin, which can be mutated to prevent beta-catenin binding to BCL9 without affecting cadherin or alpha-catenin binding. We also demonstrate that beta-catenin Y142 phosphorylation, which has been proposed to regulate BCL9-2 binding, does not directly affect the interaction of beta-catenin with either BCL9 or BCL9-2.
- Department of Biological Structure, University of Washington, Seattle, Washington 98195, USA.
Organizational Affiliation: