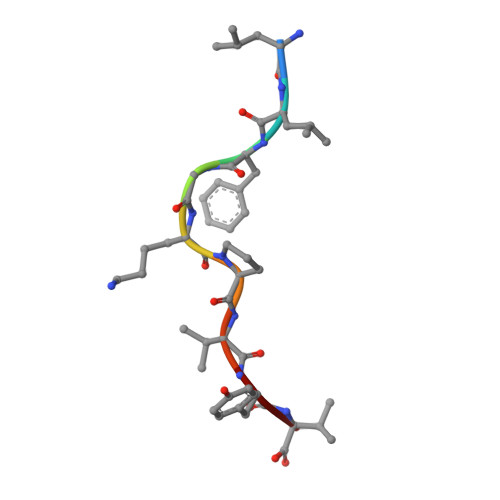
T Cell Receptor Recognition via Cooperative Conformational Plasticity.
Gagnon, S.J., Borbulevych, O.Y., Davis-Harrison, R.L., Turner, R.V., Damirjian, M., Wojnarowicz, A., Biddison, W.E., Baker, B.M.(2006) J Mol Biology 363: 228-243
- PubMed: 16962135
- DOI: https://doi.org/10.1016/j.jmb.2006.08.045
- Primary Citation of Related Structures:
2GIT, 2GJ6 - PubMed Abstract:
Although T cell receptor cross-reactivity is a fundamental property of the immune system and is implicated in numerous autoimmune pathologies, the molecular mechanisms by which T cell receptors can recognize and respond to diverse ligands are incompletely understood. In the current study we examined the response of the human T cell lymphotropic virus-1 (HTLV-1) Tax-specific T cell receptor (TCR) A6 to a panel of structurally distinct haptens coupled to the Tax 11-19 peptide with a lysine substitution at position 5 (Tax5K, LLFG[K-hapten]PVYV). The A6 TCR could cross-reactively recognize one of these haptenated peptides, Tax-5K-4-(3-Indolyl)-butyric acid (IBA), presented by HLA-A*0201. The crystal structures of Tax5K-IBA/HLA-A2 free and in complex with A6 reveal that binding is mediated by a mechanism of cooperative conformational plasticity involving conformational changes on both sides of the protein-protein interface, including the TCR complementarity determining region (CDR) loops, Valpha/Vbeta domain orientation, and the hapten-modified peptide. Our findings illustrate the complex role that protein dynamics can play in TCR cross-reactivity and highlight that T cell receptor recognition of ligand can be achieved through diverse and complex molecular mechanisms that can occur simultaneously in the interface, not limited to molecular mimicry and CDR loop shifts.
- Molecular Immunology Section, Neuroimmunology Branch, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: