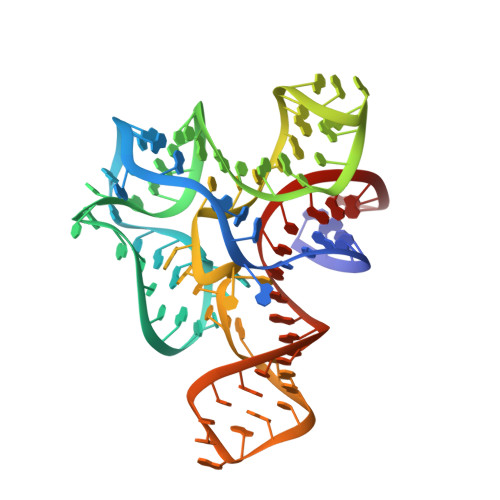
Structure of the S-adenosylmethionine riboswitch regulatory mRNA element.
Montange, R.K., Batey, R.T.(2006) Nature 441: 1172-1175
- PubMed: 16810258
- DOI: https://doi.org/10.1038/nature04819
- Primary Citation of Related Structures:
2GIS - PubMed Abstract:
Riboswitches are cis-acting genetic regulatory elements found in the 5'-untranslated regions of messenger RNAs that control gene expression through their ability to bind small molecule metabolites directly. Regulation occurs through the interplay of two domains of the RNA: an aptamer domain that responds to intracellular metabolite concentrations and an expression platform that uses two mutually exclusive secondary structures to direct a decision-making process. In Gram-positive bacteria such as Bacillus species, riboswitches control the expression of more than 2% of all genes through their ability to respond to a diverse set of metabolites including amino acids, nucleobases and protein cofactors. Here we report the 2.9-angstroms resolution crystal structure of an S-adenosylmethionine (SAM)-responsive riboswitch from Thermoanaerobacter tengcongensis complexed with S-adenosylmethionine, an RNA element that controls the expression of several genes involved in sulphur and methionine metabolism. This RNA folds into a complex three-dimensional architecture that recognizes almost every functional group of the ligand through a combination of direct and indirect readout mechanisms. Ligand binding induces the formation of a series of tertiary interactions with one of the helices, serving as a communication link between the aptamer and expression platform domains.