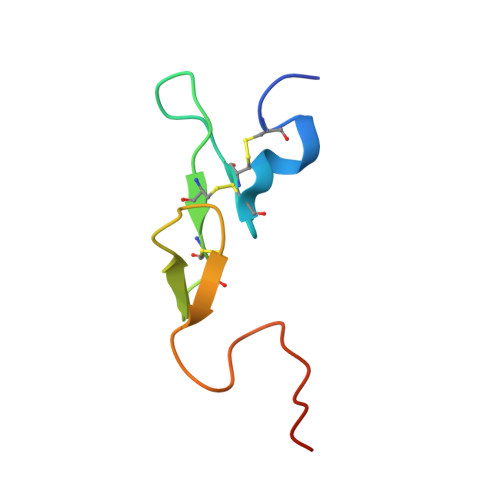
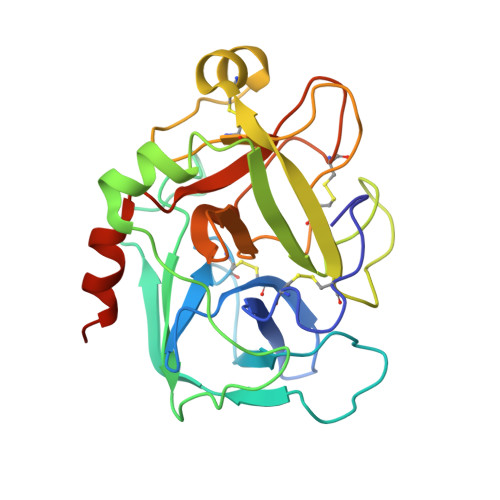
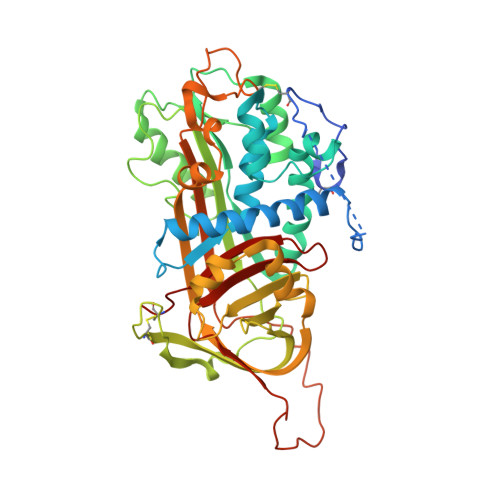
Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation.
Johnson, D.J., Li, W., Adams, T.E., Huntington, J.A.(2006) EMBO J 25: 2029-2037
- PubMed: 16619025
- DOI: https://doi.org/10.1038/sj.emboj.7601089
- Primary Citation of Related Structures:
2GD4 - PubMed Abstract:
Regulation of blood coagulation is critical for maintaining blood flow, while preventing excessive bleeding or thrombosis. One of the principal regulatory mechanisms involves heparin activation of the serpin antithrombin (AT). Inhibition of several coagulation proteases is accelerated by up to 10,000-fold by heparin, either through bridging AT and the protease or by inducing allosteric changes in the properties of AT. The anticoagulant effect of short heparin chains, including the minimal AT-specific pentasaccharide, is mediated exclusively through the allosteric activation of AT towards efficient inhibition of coagulation factors (f) IXa and Xa. Here we present the crystallographic structure of the recognition (Michaelis) complex between heparin-activated AT and S195A fXa, revealing the extensive exosite contacts that confer specificity. The heparin-induced conformational change in AT is required to allow simultaneous contacts within the active site and two distinct exosites of fXa (36-loop and the autolysis loop). This structure explains the molecular basis of protease recognition by AT, and the mechanism of action of the important therapeutic low-molecular-weight heparins.
- Department of Haematology, Division of Stuctural Medicine, Thrombosis Research Unit, Cambridge Institute for Medical Research, University of Cambridge, UK.
Organizational Affiliation: