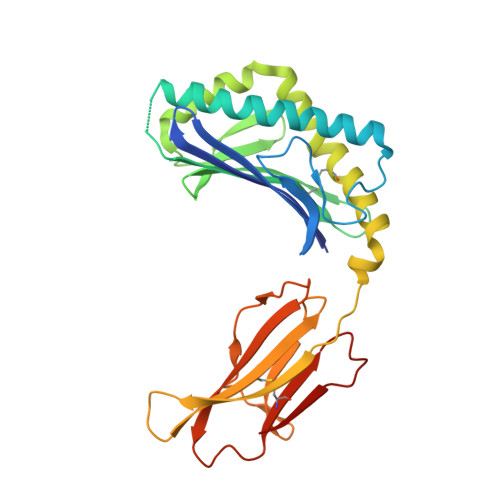
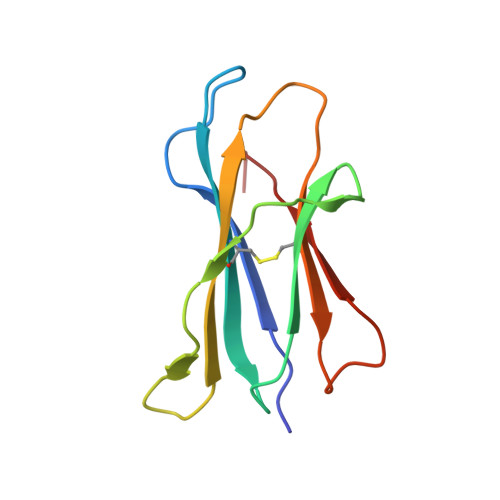
Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d.
Zajonc, D.M., Ainge, G.D., Painter, G.F., Severn, W.B., Wilson, I.A.(2006) J Immunol 177: 4577-4583
- PubMed: 16982895
- DOI: https://doi.org/10.4049/jimmunol.177.7.4577
- Primary Citation of Related Structures:
2GAZ - PubMed Abstract:
Mycobacterial phosphatidylinositol tetramannosides (PIM4) are agonists for a distinct population of invariant human (Valpha24) and mouse (Valpha14) NKT cells, when presented by CD1d. We determined the crystal structure at 2.6-A resolution of mouse CD1d bound to a synthetic dipalmitoyl-PIM2. Natural PIM2, which differs in its fatty acid composition is a biosynthetic precursor of PIM4, PIM6, lipomannan, and lipoarabinomannan. The PIM2 headgroup (inositol-dimannoside) is the most complex to date among all the crystallized CD1d ligands and is remarkably ordered in the CD1d binding groove. A specific hydrogen-bonding network between PIM2 and CD1d orients the headgroup in the center of the binding groove and above the A' pocket. A central cluster of hydrophilic CD1d residues (Asp(153), Thr(156), Ser(76), Arg(79)) interacts with the phosphate, inositol, and alpha1-alpha6-linked mannose of the headgroup, whereas additional specificity for the alpha1- and alpha2-linked mannose is conferred by Thr(159). The additional two mannoses in PIM4, relative to PIM2, are located at the distal 6' carbon of the alpha1-alpha6-linked mannose and would project away from the CD1d binding groove for interaction with the TCR. Compared with other CD1d-sphingolipid structures, PIM2 has an increased number of polar interactions between its headgroup and CD1, but reduced specificity for the diacylglycerol backbone. Thus, novel NKT cell agonists can be designed that focus on substitutions of the headgroup rather than on reducing lipid chain length, as in OCH and PBS-25, two potent variants of the highly stimulatory invariant NKT cell agonist alpha-galactosylceramide.
- Department of Molecular Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: