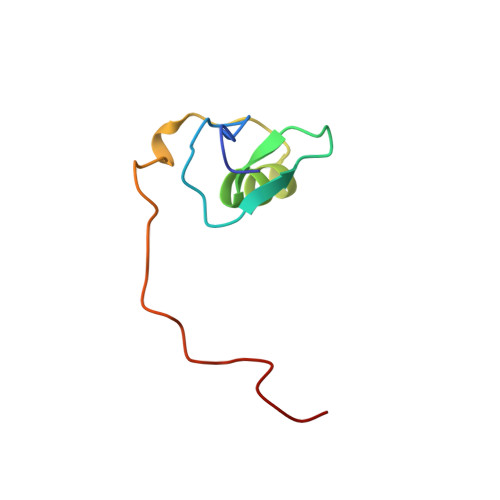
Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Tjandra, N., Omichinski, J.G., Gronenborn, A.M., Clore, G.M., Bax, A.(1997) Nat Struct Biol 4: 732-738
- PubMed: 9303001
- DOI: https://doi.org/10.1038/nsb0997-732
- Primary Citation of Related Structures:
2GAT, 3GAT - PubMed Abstract:
Anisotropy of the molecular magnetic susceptibility gives rise to a small degree of alignment. The resulting residual dipolar couplings, which can now be measured with the advent of higher magnetic fields in NMR, contain information on the orientation of the internuclear vectors relative to the molecular magnetic susceptibility tensor, thereby providing information on long range order that is not accessible by any of the solution NMR parameters currently used in structure determination. Thus, the dipolar couplings constitute unique and powerful restraints in determining the structures of magnetically oriented macromolecules in solution. The method is demonstrated on a complex of the DNA-binding domain of the transcription factor GATA-1 with a 16 base pair oligodeoxyribonucleotide.
- Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892-0520, USA.
Organizational Affiliation: