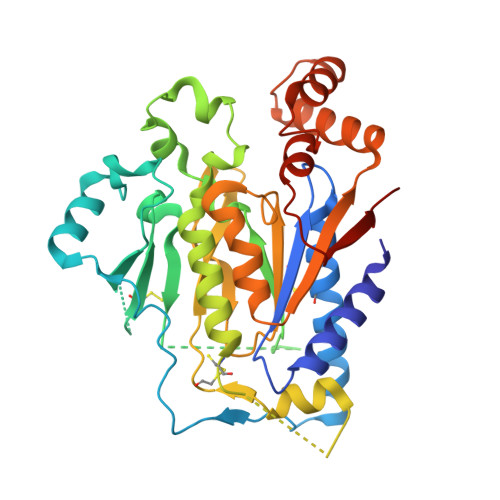
Crystal structure and functional analysis identify the P-loop containing protein YFH7 of Saccharomyces cerevisiae as an ATP-dependent kinase.
Gueguen-Chaignon, V., Chaptal, V., Lariviere, L., Costa, N., Lopes, P., Morera, S., Nessler, S.(2007) Proteins 71: 804-812
- PubMed: 18004758
- DOI: https://doi.org/10.1002/prot.21740
- Primary Citation of Related Structures:
2GA8, 2GAA - PubMed Abstract:
Genome sequencing projects have revealed that P-loop proteins are highly represented in all organisms and that many of them have no attributed function. They are characterized by a conserved nucleotide-binding domain and carry different activities implicated in many cellular processes. Saccharomyces cerevisiae YFH7 is one of these P-loop proteins of unknown function. In this work we tried to integrate bioinformatics, structure, and enzymology to discover the function of YFH7. Sequence analysis revealed that yeast YFH7 is a yeast-specific protein showing weak similarity with the phosphoribulokinase/uridine kinase/bacterial pantothenate kinase (PRK/URK/PANK) subfamily of P-loop containing kinases. A large insertion of about 100 residues distinguishes YFH7 from other members of the family. The 1.95 A resolution crystal structure of YFH7 solved using the SAD method confirmed that YFH7 has a fold similar to the PRK/URK/PANK family, with the characteristic core, lid, and NMP(bind) domains. An additional alpha/beta domain of novel topology corresponds to the large sequence insertion. Structural and ligand binding analysis combined with enzymatic assays suggest that YFH7 is an ATP-dependent small molecule kinase with new substrate specificity.
- Laboratoire d'Enzymologie et Biochimie Structurales, CNRS, 1 av. de la terrasse, 91198 Gif-sur Yvette, France.
Organizational Affiliation: