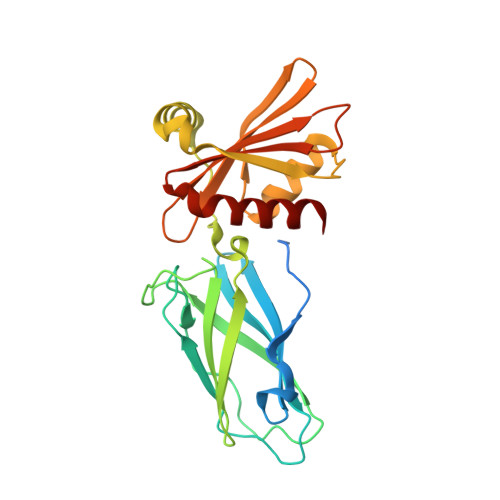
Molecular Switches Involving the AP-2 beta2 Appendage Regulate Endocytic Cargo Selection and Clathrin Coat Assembly
Edeling, M.A., Mishra, S.K., Keyel, P.A., Steinhauser, A.L., Collins, B.M., Roth, R., Heuser, J.E., Owen, D.J., Traub, L.M.(2006) Dev Cell 10: 329-342
- PubMed: 16516836
- DOI: https://doi.org/10.1016/j.devcel.2006.01.016
- Primary Citation of Related Structures:
2G30 - PubMed Abstract:
Clathrin-associated sorting proteins (CLASPs) expand the repertoire of endocytic cargo sorted into clathrin-coated vesicles beyond the transmembrane proteins that bind physically to the AP-2 adaptor. LDL and GPCRs are internalized by ARH and beta-arrestin, respectively. We show that these two CLASPs bind selectively to the AP-2 beta2 appendage platform via an alpha-helical [DE](n)X(1-2)FXX[FL]XXXR motif, and that this motif also occurs and is functional in the epsins. In beta-arrestin, this motif maintains the endocytosis-incompetent state by binding back on the folded core of the protein in a beta strand conformation. Triggered via a beta-arrestin/GPCR interaction, the motif must be displaced and must undergo a strand to helix transition to enable the beta2 appendage binding that drives GPCR-beta-arrestin complexes into clathrin coats. Another interaction surface on the beta2 appendage sandwich is identified for proteins such as eps15 and clathrin, suggesting a mechanism by which clathrin displaces eps15 to lattice edges during assembly.
- Cambridge Institute for Medical Research, University of Cambridge, Cambridge CB2 2XY, United Kingdom.
Organizational Affiliation: