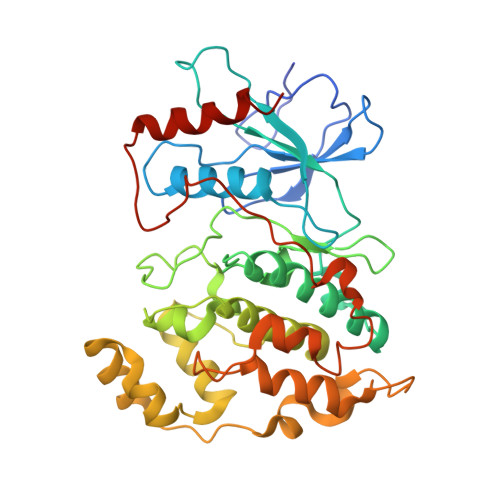
Synthesis and SAR of 1,9-dihydro-9-hydroxypyrazolo[3,4-b]quinolin-4-ones as novel, selective c-Jun N-terminal kinase inhibitors.
Liu, M., Xin, Z., Clampit, J.E., Wang, S., Gum, R.J., Haasch, D.L., Trevillyan, J.M., Abad-Zapatero, C., Fry, E.H., Sham, H.L., Liu, G.(2006) Bioorg Med Chem Lett 16: 2590-2594
- PubMed: 16527482
- DOI: https://doi.org/10.1016/j.bmcl.2006.02.046
- Primary Citation of Related Structures:
2G01 - PubMed Abstract:
A novel class of 1,9-dihydro-9-hydroxypyrazolo[3,4-b]quinolin-4-ones as c-Jun-N-terminal kinase (JNK) inhibitors is described. These compounds were synthesized via the condensation of 2-nitrobenzaldehydes and hydroxypyrazoles. The structure-activity relationships (SAR) and kinase selectivity profile of the inhibitors are also discussed. Compound 16 was identified as a potent JNK inhibitor with good cellular potency.
- Metabolic Disease Research, Global Pharmaceutical Research and Development, Abbott Laboratories, 100 Abbott Park Road, Abbott Park, IL 60064-6098, USA. mei.liu@abbott.com
Organizational Affiliation: