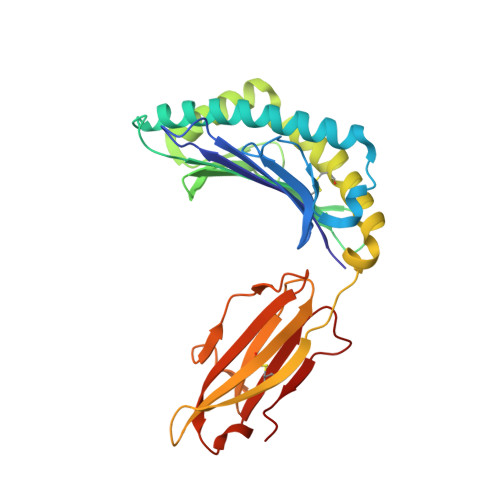
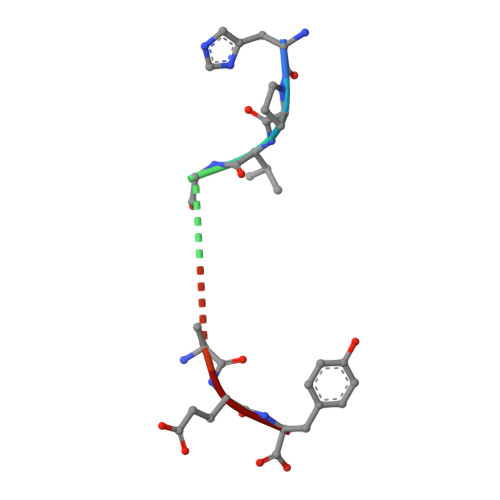
TCR alpha genes direct MHC restriction in the potent human T cell response to a class I-bound viral epitope.
Miles, J.J., Borg, N.A., Brennan, R.M., Tynan, F.E., Kjer-Nielsen, L., Silins, S.L., Bell, M.J., Burrows, J.M., McCluskey, J., Rossjohn, J., Burrows, S.R.(2006) J Immunol 177: 6804-6814
- PubMed: 17082594
- DOI: https://doi.org/10.4049/jimmunol.177.10.6804
- Primary Citation of Related Structures:
2FYY, 2FZ3 - PubMed Abstract:
The underlying generic properties of alphabeta TCRs that control MHC restriction remain largely unresolved. To investigate MHC restriction, we have examined the CTL response to a viral epitope that binds promiscuously to two human leukocyte Ags (HLAs) that differ by a single amino acid at position 156. Individuals expressing either HLA-B*3501 (156Leucine) or HLA-B*3508 (156Arginine) showed a potent CTL response to the 407HPVGEADYFEY417 epitope from EBV. Interestingly, the response was characterized by highly restricted TCR beta-chain usage in both HLA-B*3501+ and HLA-B*3508+ individuals; however, this conserved TRBV9+ beta-chain was associated with distinct TCR alpha-chains depending upon the HLA-B*35 allele expressed by the virus-exposed host. Functional assays confirmed that TCR alpha-chain usage determined the HLA restriction of the CTLs. Structural studies revealed significant differences in the mobility of the peptide when bound to HLA-B*3501 or HLA-B*3508. In HLA-B*3501, the bulged section of the peptide was disordered, whereas in HLA-B*3508 the bulged epitope adopted an ordered conformation. Collectively, these data demonstrate not only that mobile MHC-bound peptides can be highly immunogenic but can also stimulate an extremely biased TCR repertoire. In addition, TCR alpha-chain usage is shown to play a critical role in controlling MHC restriction between closely related allomorphs.
- Cellular Immunology Laboratory, Queensland Institute of Medical Research, Brisbane, Australia.
Organizational Affiliation: