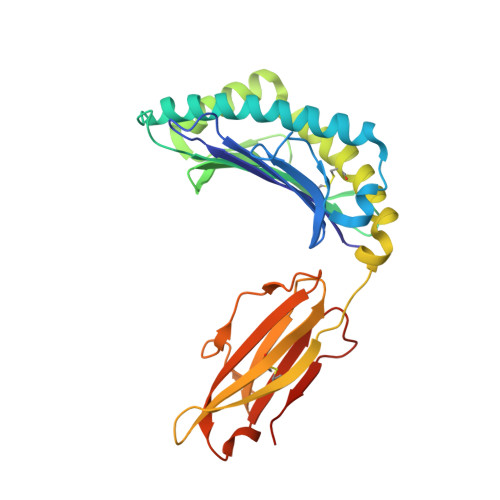
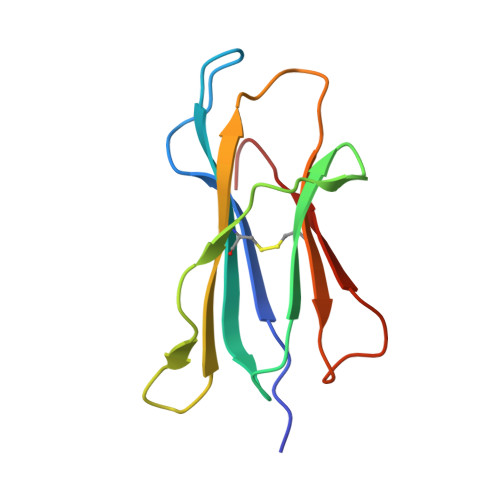
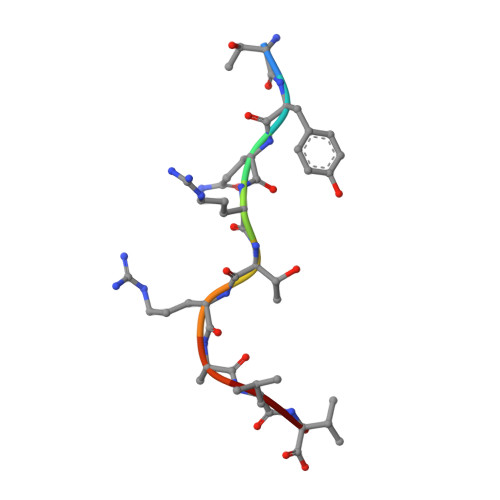
Structural Definition of the H-2Kd Peptide-binding Motif.
Mitaksov, V., Fremont, D.H.(2006) J Biological Chem 281: 10618-10625
- PubMed: 16473882
- DOI: https://doi.org/10.1074/jbc.M510511200
- Primary Citation of Related Structures:
2FWO - PubMed Abstract:
Classic major histocompatibility complex (MHC) proteins associate with antigen- and self-derived peptides in an allele-specific manner. Herein we present the crystal structure of the MHC class I protein H-2K(d) (K(d)) expressed by BALB/c mice in complex with an antigenic peptide derived from influenza A/PR/8/34 nucleoprotein (Flu, residues 147-155, TYQRTRALV). Analysis of our structure in conjunction with the sequences of naturally processed epitopes provides a comprehensive understanding of the dominant K(d) peptide-binding motif. We find that Flu residues Tyr(P2), Thr(P5), and Val(P9) are sequestered into the B, C, and F pockets of the K(d) groove, respectively. The shape and chemistry of the polymorphic B pocket make it an optimal binding site for the side chain of Tyr(P2) as the dominant anchoring residue of nonameric peptides. The non-polar F pocket limits the amino acid repertoire at P9 to hydrophobic residues such as Ile, Leu, or Val, whereas the C pocket restricts the size of the P5-anchoring side chain. We also show that Flu is accommodated in the complex through an unfavorable kink in the otherwise extended peptide backbone due to the presence of a prominent ridge in the K(d) groove. Surprisingly, this backbone conformation is strikingly similar to D(b)-presented peptides despite the fact that these proteins employ distinct motif-anchoring strategies. The results presented in this study provide a solid foundation for the understanding of K(d)-restricted antigen presentation and recognition events.
- Department of Pathology and Immunology, Washington University School of Medicine, 660 South Euclid Avenue, St. Louis, MO 63110, USA.
Organizational Affiliation: