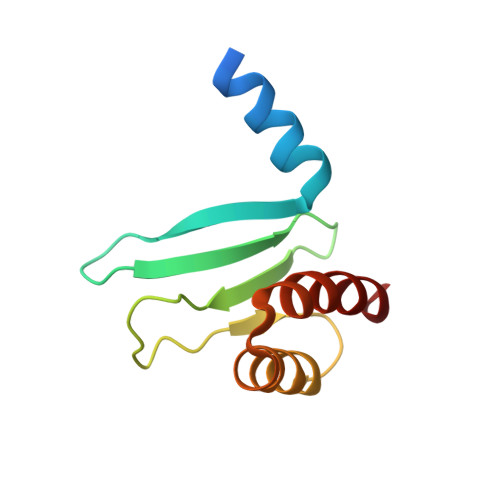
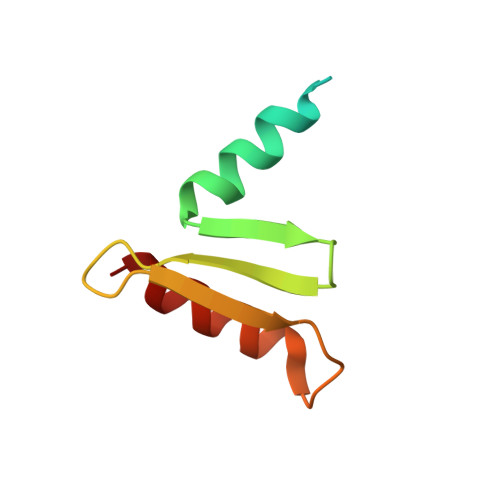
Structure of a central component of the yeast kinetochore: the spc24p/spc25p globular domain.
Wei, R.R., Schnell, J.R., Larsen, N.A., Sorger, P.K., Chou, J.J., Harrison, S.C.(2006) Structure 6: 1003-1009
- PubMed: 16765893
- DOI: https://doi.org/10.1016/j.str.2006.04.007
- Primary Citation of Related Structures:
2FV4 - PubMed Abstract:
The Ndc80 complex, a kinetochore component conserved from yeast to humans, is essential for proper chromosome alignment and segregation during mitosis. It is an approximately 570 A long, rod-shaped assembly of four proteins--Ndc80p (Hec1), Nuf2p, Spc24p, and Spc25p--with globular regions at either end of a central shaft. The complex bridges from the centromere-proximal inner kinetochore layer at its Spc24/Spc25 globular end to the microtubule binding outer kinetochore layer at its Ndc80/Nuf2 globular end. We report the atomic structures of the Spc24/Spc25 globular domain, determined both by X-ray crystallography at 1.9 A resolution and by NMR. Spc24 and Spc25 fold tightly together into a single globular entity with pseudo-2-fold symmetry. Conserved residues line a common hydrophobic core and the bottom of a cleft, indicating that the functional orthologs from other eukaryotes will have the same structure and suggesting a docking site for components of the inner kinetochore.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts 02115, USA.
Organizational Affiliation: