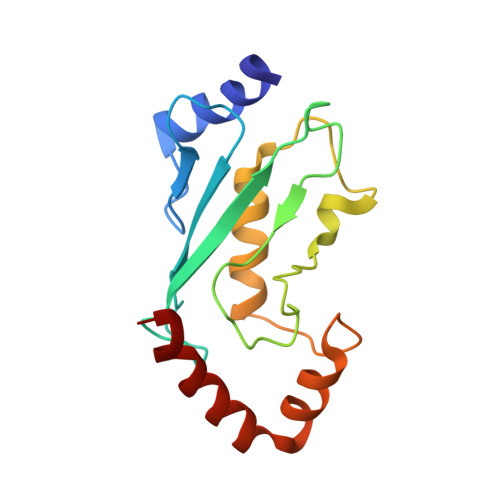
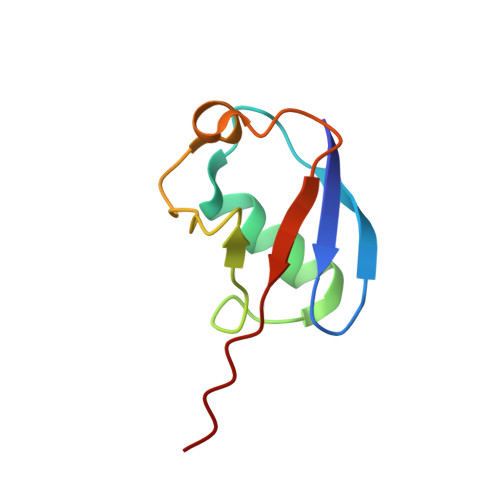
A UbcH5/Ubiquitin Noncovalent Complex Is Required for Processive BRCA1-Directed Ubiquitination.
Brzovic, P.S., Lissounov, A., Christensen, D.E., Hoyt, D.W., Klevit, R.E.(2006) Mol Cell 21: 873-880
- PubMed: 16543155
- DOI: https://doi.org/10.1016/j.molcel.2006.02.008
- Primary Citation of Related Structures:
2FUH - PubMed Abstract:
Protein ubiquitination is a powerful regulatory modification that influences nearly every aspect of eukaryotic cell biology. The general pathway for ubiquitin (Ub) modification requires the sequential activities of a Ub-activating enzyme (E1), a Ub transfer enzyme (E2), and a Ub ligase (E3). The E2 must recognize both the E1 and a cognate E3 in addition to carrying activated Ub. These central functions are performed by a topologically conserved alpha/beta-fold core domain of approximately 150 residues shared by all E2s. However, as presented herein, the UbcH5 family of E2s can also bind Ub noncovalently on a surface well removed from the E2 active site. We present the solution structure of the UbcH5c/Ub noncovalent complex and demonstrate that this noncovalent interaction permits self-assembly of activated UbcH5c approximately Ub molecules. Self-assembly has profound consequences for the processive formation of polyubiquitin (poly-Ub) chains in ubiquitination reactions directed by the breast and ovarian cancer tumor susceptibility protein BRCA1.
- Department of Biochemistry, University of Washington, Seattle, 98195, USA.
Organizational Affiliation: