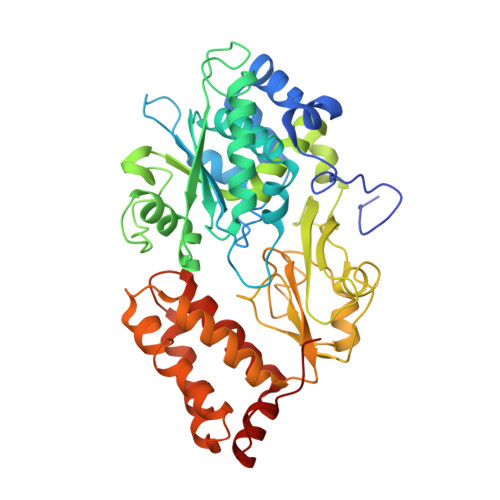
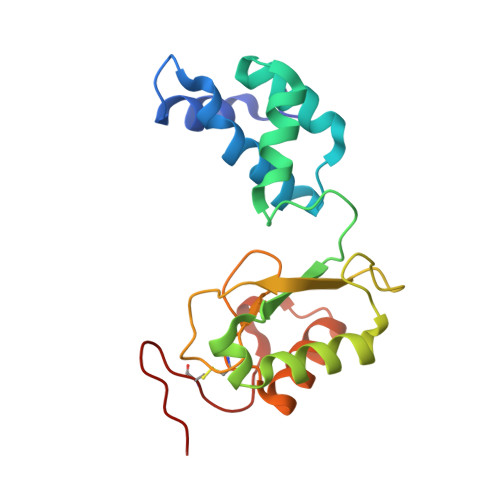
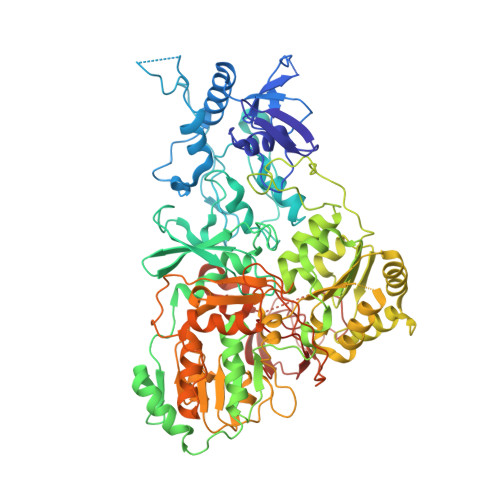
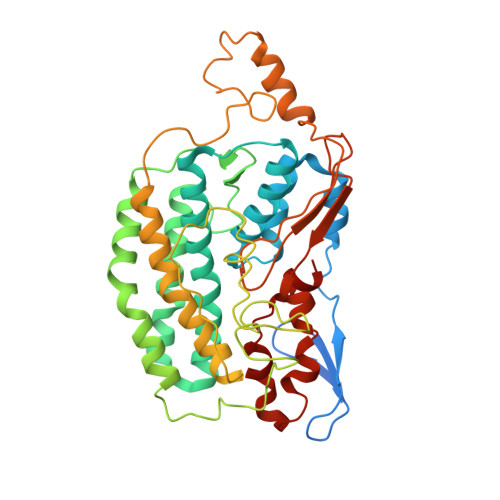
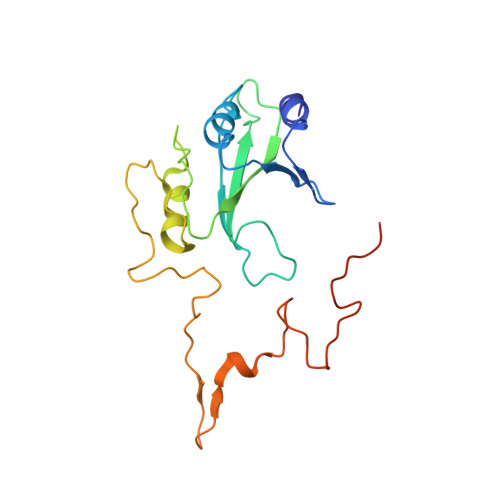
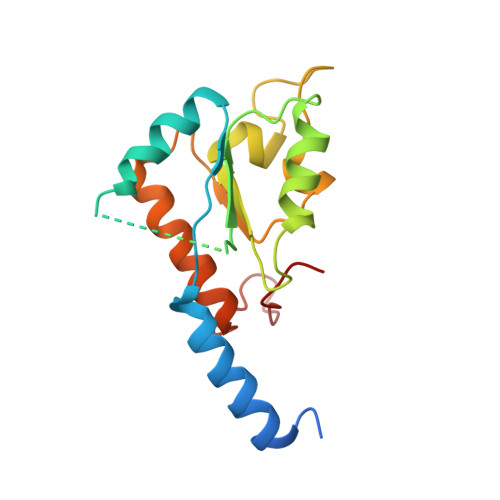
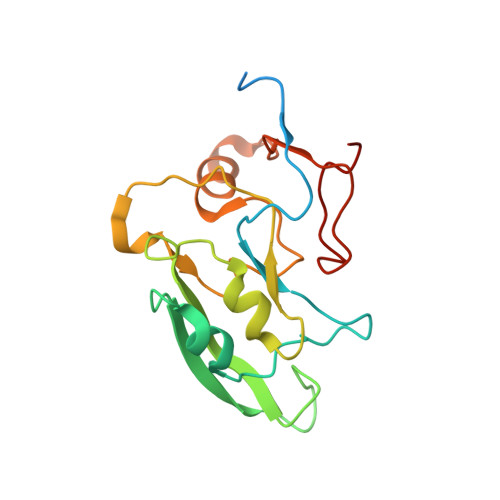
Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus.
Sazanov, L.A., Hinchliffe, P.(2006) Science 311: 1430-1436
- PubMed: 16469879
- DOI: https://doi.org/10.1126/science.1123809
- Primary Citation of Related Structures:
2FUG - PubMed Abstract:
Respiratory complex I plays a central role in cellular energy production in bacteria and mitochondria. Its dysfunction is implicated in many human neurodegenerative diseases, as well as in aging. The crystal structure of the hydrophilic domain (peripheral arm) of complex I from Thermus thermophilus has been solved at 3.3 angstrom resolution. This subcomplex consists of eight subunits and contains all the redox centers of the enzyme, including nine iron-sulfur clusters. The primary electron acceptor, flavin-mononucleotide, is within electron transfer distance of cluster N3, leading to the main redox pathway, and of the distal cluster N1a, a possible antioxidant. The structure reveals new aspects of the mechanism and evolution of the enzyme. The terminal cluster N2 is coordinated, uniquely, by two consecutive cysteines. The novel subunit Nqo15 has a similar fold to the mitochondrial iron chaperone frataxin, and it may be involved in iron-sulfur cluster regeneration in the complex.
- Medical Research Council Dunn Human Nutrition Unit, Wellcome Trust/MRC Building, Hills Road, Cambridge CB2 2XY, U.K. sazanov@mrc-dunn.cam.ac.uk
Organizational Affiliation: