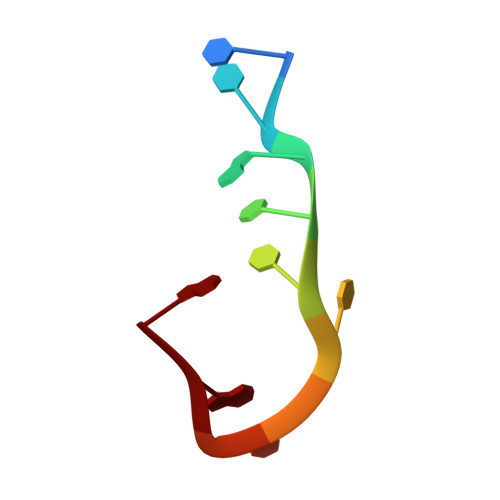
Impact of DNA hairpin folding energetics on antibody-ssDNA association.
Ou, Z., Bottoms, C.A., Henzl, M.T., Tanner, J.J.(2007) J Mol Biology 374: 1029-1040
- PubMed: 18028946
- DOI: https://doi.org/10.1016/j.jmb.2007.09.084
- Primary Citation of Related Structures:
2FR4 - PubMed Abstract:
Deposition of anti-DNA antibodies in the kidney contributes to the pathogenesis of the autoimmune disease, systemic lupus erythematosus. Antibodies that bind to hairpin-forming DNA ligands may be particularly prone to deposition. Here we report the first structure of a Fab complexed with hairpin-forming DNA. The ligand used for co-crystallization is 5'-d [CTG(CCTT)CAG]-3', which has a predicted hairpin structure consisting of a four-nucleotide loop (CCTT) and a stem of three base-pairs. The 1.95 A resolution crystal structure of Fab DNA-1 complexed with this ligand shows that the conformation of the bound ligand differs radically from the predicted hairpin conformation. The three base-pairs in the stem are absent in the bound form. The protein binds to the last six nucleotides at the 3' end of the ligand. These nucleotides form a loop (TTCA) closed by a G:C base-pair in the bound state. Stacking of aromatic side-chains against DNA bases is the dominant interaction in the complex. Interactions with the DNA backbone are conspicuously absent. Thermodynamics of binding are examined using isothermal titration calorimetry. The apparent dissociation constant is 4 microM, and binding is enthalpically favorable and entropically unfavorable. Increasing the number of base-pairs in the DNA stem from three to six decreases binding affinity. These data suggest a conformational selection binding mechanism in which the Fab binds preferentially to the unstructured state of the ligand. In this interpretation, the ligand binding and ligand folding equilibria are coupled, with lower hairpin stability leading to greater effective binding affinity. Thus, pre-organization of the DNA loop into the preferred binding conformation does not play a major role in complexation. Rather, it is argued that the stem of the hairpin serves to reduce the degrees of freedom in the free DNA ligand, thereby limiting the entropic cost attendant to complexation with the Fab.
- Department of Biochemistry, University of Missouri-Columbia, Columbia, MO 65211, USA.
Organizational Affiliation: