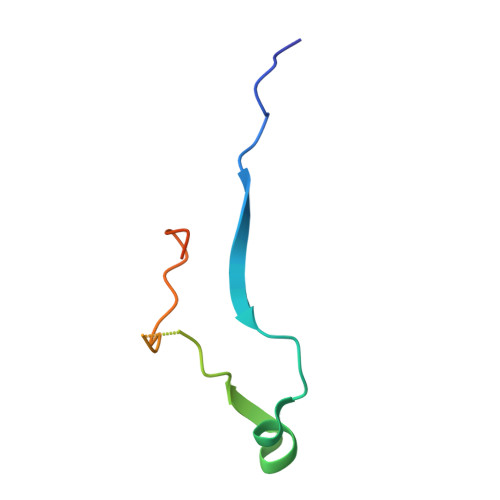
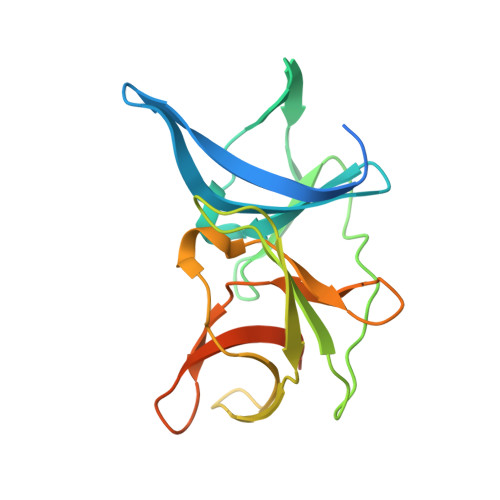
Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus.
Erbel, P., Schiering, N., D'Arcy, A., Renatus, M., Kroemer, M., Lim, S.P., Yin, Z., Keller, T.H., Vasudevan, S.G., Hommel, U.(2006) Nat Struct Mol Biol 13: 372-373
- PubMed: 16532006
- DOI: https://doi.org/10.1038/nsmb1073
- Primary Citation of Related Structures:
2FOM, 2FP7 - PubMed Abstract:
The replication of flaviviruses requires the correct processing of their polyprotein by the viral NS3 protease (NS3pro). Essential for the activation of NS3pro is a 47-residue region of NS2B. Here we report the crystal structures of a dengue NS2B-NS3pro complex and a West Nile virus NS2B-NS3pro complex with a substrate-based inhibitor. These structures identify key residues for NS3pro substrate recognition and clarify the mechanism of NS3pro activation.
- Novartis Institutes for Biomedical Research, Protease Platform, 4002 Basel, Switzerland.
Organizational Affiliation: