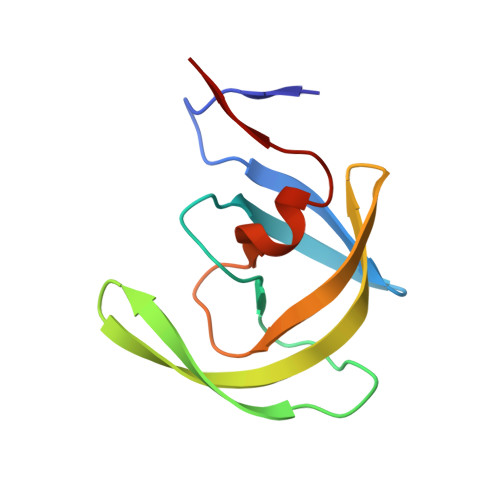
Mechanism of substrate recognition by drug-resistant human immunodeficiency virus type 1 protease variants revealed by a novel structural intermediate.
Prabu-Jeyabalan, M., Nalivaika, E.A., Romano, K., Schiffer, C.A.(2006) J Virol 80: 3607-3616
- PubMed: 16537628
- DOI: https://doi.org/10.1128/JVI.80.7.3607-3616.2006
- Primary Citation of Related Structures:
2FNS, 2FNT - PubMed Abstract:
Human immunodeficiency virus type 1 (HIV-1) protease processes and cleaves the Gag and Gag-Pol polyproteins, allowing viral maturation, and therefore is an important target for antiviral therapy. Ligand binding occurs when the flaps open, allowing access to the active site. This flexibility in flap geometry makes trapping and crystallizing structural intermediates in substrate binding challenging. In this study, we report two crystal structures of two HIV-1 protease variants bound with their corresponding nucleocapsid-p1 variant. One of the flaps in each of these structures exhibits an unusual "intermediate" conformation. Analysis of the flap-intermediate and flap-closed crystal structures reveals that the intermonomer flap movements may be asynchronous and that the flap which wraps over the P3 to P1 (P3-P1) residues of the substrate might close first. This is consistent with our hypothesis that the P3-P1 region is crucial for substrate recognition. The intermediate conformation is conserved in both the wild-type and drug-resistant variants. The structural differences between the variants are evident only when the flaps are closed. Thus, a plausible structural model for the adaptability of HIV-1 protease to recognize substrates in the presence of drug-resistant mutations has been proposed.
- Department of Biochemistry & Molecular Pharmacology, University of Massachusetts Medical School, 364 Plantation St., Worcester, MA 01605, USA.
Organizational Affiliation: