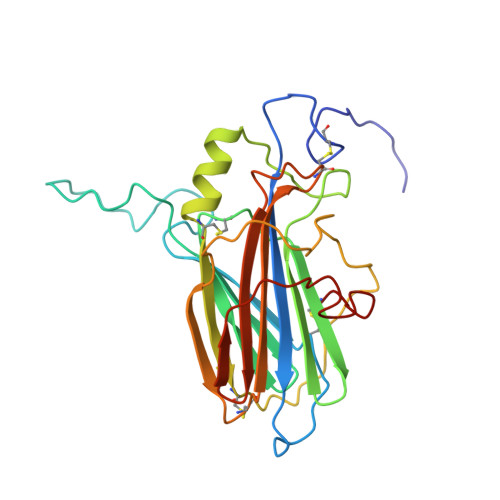
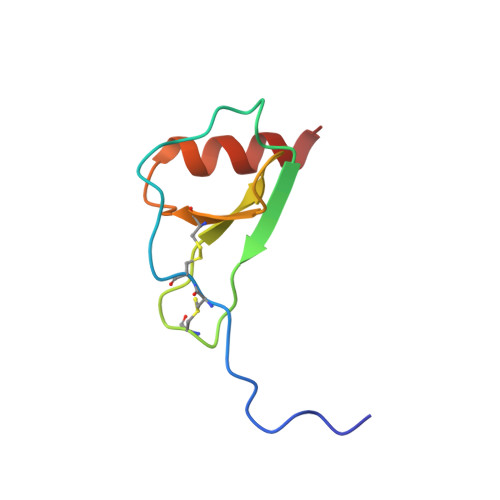
Solution structure of the complex between poxvirus-encoded CC chemokine inhibitor vCCI and human MIP-1beta.
Zhang, L., Derider, M., McCornack, M.A., Jao, S.C., Isern, N., Ness, T., Moyer, R., Liwang, P.J.(2006) Proc Natl Acad Sci U S A 103: 13985-13990
- PubMed: 16963564
- DOI: https://doi.org/10.1073/pnas.0602142103
- Primary Citation of Related Structures:
2FFK, 2FIN - PubMed Abstract:
Chemokines (chemotactic cytokines) comprise a large family of proteins that recruit and activate leukocytes, giving chemokines a major role in both immune response and inflammation-related diseases. The poxvirus-encoded viral CC chemokine inhibitor (vCCI) binds to many CC chemokines with high affinity, acting as a potent inhibitor of chemokine action. We have used heteronuclear multidimensional NMR to determine the structure of an orthopoxvirus vCCI in complex with a human CC chemokine, MIP-1beta (macrophage inflammatory protein 1beta). vCCI binds to the chemokine with 1:1 stoichiometry, forming a complex of 311 aa. vCCI uses residues from its beta-sheet II to interact with a surface of MIP-1beta that includes residues adjacent to its N terminus, as well as residues in the 20's region and the 40's loop. This structure reveals the strategy used by vCCI to tightly bind numerous chemokines while retaining selectivity for the CC chemokine subfamily.
- Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843-2128, USA.
Organizational Affiliation: