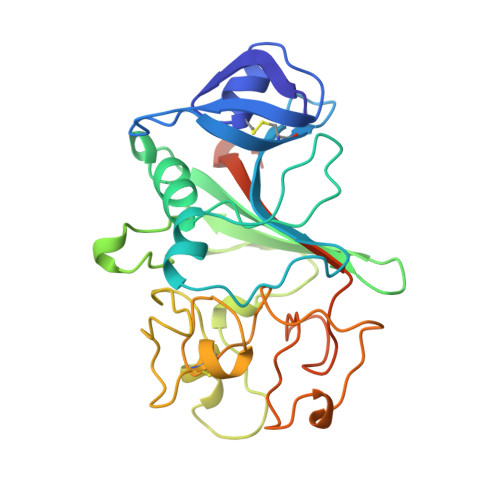
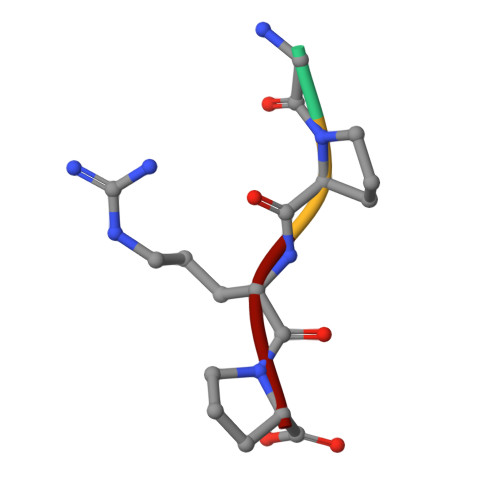
The primary fibrin polymerization pocket: three-dimensional structure of a 30-kDa C-terminal gamma chain fragment complexed with the peptide Gly-Pro-Arg-Pro.
Pratt, K.P., Cote, H.C., Chung, D.W., Stenkamp, R.E., Davie, E.W.(1997) Proc Natl Acad Sci U S A 94: 7176-7181
- PubMed: 9207064
- DOI: https://doi.org/10.1073/pnas.94.14.7176
- Primary Citation of Related Structures:
2FIB, 3FIB - PubMed Abstract:
After vascular injury, a cascade of serine protease activations leads to the conversion of the soluble fibrinogen molecule into fibrin. The fibrin monomers then polymerize spontaneously and noncovalently to form a fibrin gel. The primary interaction of this polymerization reaction is between the newly exposed N-terminal Gly-Pro-Arg sequence of the alpha chain of one fibrin molecule and the C-terminal region of a gamma chain of an adjacent fibrin(ogen) molecule. In this report, the polymerization pocket has been identified by determining the crystal structure of a 30-kDa C-terminal fragment of the fibrin(ogen) gamma chain complexed with the peptide Gly-Pro-Arg-Pro. This peptide mimics the N terminus of the alpha chain of fibrin. The conformational change in the protein upon binding the peptide is subtle, with electrostatic interactions primarily mediating the association. This is consistent with biophysical experiments carried out over the last 50 years on this fundamental polymerization reaction.
- Department of Biochemistry, University of Washington, Seattle, WA 98195, USA. kpratt@u.washington.edu
Organizational Affiliation: