Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain
Olsen, S.K., Li, J.Y.H., Bromleigh, C., Eliseenkova, A.V., Ibrahimi, O.A., Lao, Z., Zhang, F., Linhardt, R.J., Joyner, A.L., Mohammadi, M.(2006) Genes Dev 20: 185-198
- PubMed: 16384934
- DOI: https://doi.org/10.1101/gad.1365406
- Primary Citation of Related Structures:
2FDB - PubMed Abstract:
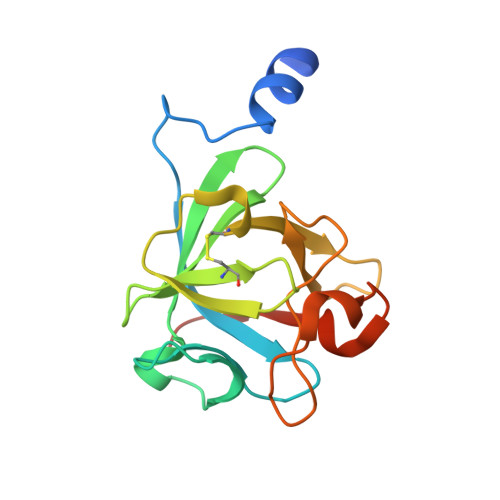
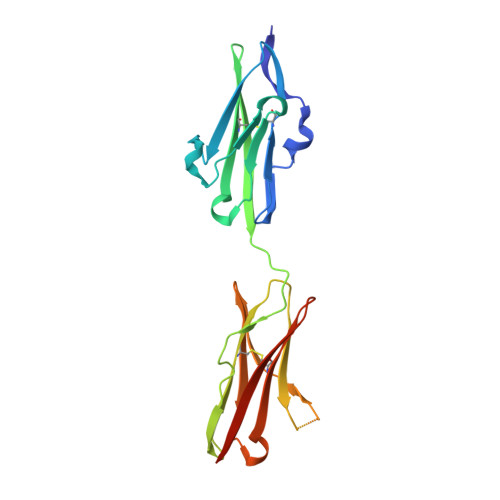
Two of the four human FGF8 splice isoforms, FGF8a and FGF8b, are expressed in the mid-hindbrain region during development. Although the only difference between these isoforms is the presence of an additional 11 amino acids at the N terminus of FGF8b, these isoforms possess remarkably different abilities to pattern the midbrain and anterior hindbrain. To reveal the structural basis by which alternative splicing modulates the organizing activity of FGF8, we solved the crystal structure of FGF8b in complex with the "c" splice isoform of FGF receptor 2 (FGFR2c). Using surface plasmon resonance (SPR), we also characterized the receptor-binding specificity of FGF8a and FGF8b, the "b" isoform of FGF17 (FGF17b), and FGF18. The FGF8b-FGFR2c structure shows that alternative splicing permits a single additional contact between phenylalanine 32 (F32) of FGF8b and a hydrophobic groove within Ig domain 3 of the receptor that is also present in FGFR1c, FGFR3c, and FGFR4. Consistent with the structure, mutation of F32 to alanine reduces the affinity of FGF8b toward all these receptors to levels characteristic of FGF8a. More importantly, analysis of the mid-hindbrain patterning ability of the FGF8b(F32A) mutant in chick embryos and murine midbrain explants shows that this mutation functionally converts FGF8b to FGF8a. Moreover, our data suggest that the intermediate receptor-binding affinities of FGF17b and FGF18, relative to FGF8a and FGF8b, also account for the distinct patterning abilities of these two ligands. We also show that the mode of FGF8 receptor-binding specificity is distinct from that of other FGFs and provide the first biochemical evidence for a physiological FGF8b-FGFR1c interaction during mid-hindbrain development. Consistent with the indispensable role of FGF8 in embryonic development, we show that the FGF8 mode of receptor binding appeared as early as in nematodes and has been preserved throughout evolution.
- Department of Pharmacology, New York University School of Medicine, New York, New York 10016, USA.
Organizational Affiliation: